Random walks on mutual microRNA-target gene interaction network improve the prediction of disease-associated microRNAs
- PMID: 29137601
- PMCID: PMC5686822
- DOI: 10.1186/s12859-017-1924-1
Random walks on mutual microRNA-target gene interaction network improve the prediction of disease-associated microRNAs
Abstract
Background: MicroRNAs (miRNAs) have been shown to play an important role in pathological initiation, progression and maintenance. Because identification in the laboratory of disease-related miRNAs is not straightforward, numerous network-based methods have been developed to predict novel miRNAs in silico. Homogeneous networks (in which every node is a miRNA) based on the targets shared between miRNAs have been widely used to predict their role in disease phenotypes. Although such homogeneous networks can predict potential disease-associated miRNAs, they do not consider the roles of the target genes of the miRNAs. Here, we introduce a novel method based on a heterogeneous network that not only considers miRNAs but also the corresponding target genes in the network model.
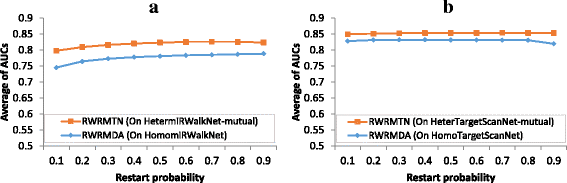
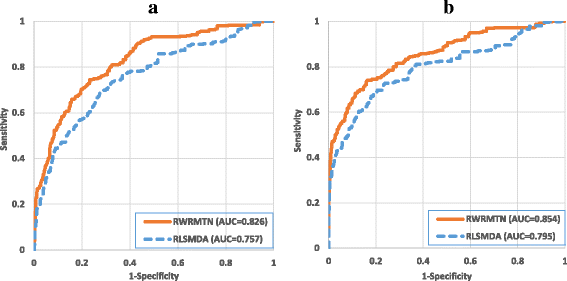
Results: Instead of constructing homogeneous miRNA networks, we built heterogeneous miRNA networks consisting of both miRNAs and their target genes, using databases of known miRNA-target gene interactions. In addition, as recent studies demonstrated reciprocal regulatory relations between miRNAs and their target genes, we considered these heterogeneous miRNA networks to be undirected, assuming mutual miRNA-target interactions. Next, we introduced a novel method (RWRMTN) operating on these mutual heterogeneous miRNA networks to rank candidate disease-related miRNAs using a random walk with restart (RWR) based algorithm. Using both known disease-associated miRNAs and their target genes as seed nodes, the method can identify additional miRNAs involved in the disease phenotype. Experiments indicated that RWRMTN outperformed two existing state-of-the-art methods: RWRMDA, a network-based method that also uses a RWR on homogeneous (rather than heterogeneous) miRNA networks, and RLSMDA, a machine learning-based method. Interestingly, we could relate this performance gain to the emergence of "disease modules" in the heterogeneous miRNA networks used as input for the algorithm. Moreover, we could demonstrate that RWRMTN is stable, performing well when using both experimentally validated and predicted miRNA-target gene interaction data for network construction. Finally, using RWRMTN, we identified 76 novel miRNAs associated with 23 disease phenotypes which were present in a recent database of known disease-miRNA associations.
Conclusions: Summarizing, using random walks on mutual miRNA-target networks improves the prediction of novel disease-associated miRNAs because of the existence of "disease modules" in these networks.
Keywords: Disease-associated microRNAs; Network analysis; Random walk with restart; microRNA targets.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

