The secreted neurotrophin Spätzle 3 promotes glial morphogenesis and supports neuronal survival and function
- PMID: 29138279
- PMCID: PMC5733495
- DOI: 10.1101/gad.305888.117
The secreted neurotrophin Spätzle 3 promotes glial morphogenesis and supports neuronal survival and function
Abstract
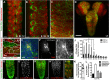
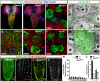
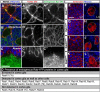
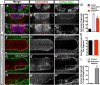
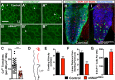
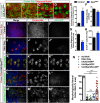
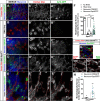
Most glial functions depend on establishing intimate morphological relationships with neurons. Significant progress has been made in understanding neuron-glia signaling at synaptic and axonal contacts, but how glia support neuronal cell bodies is unclear. Here we explored the growth and functions of Drosophila cortex glia (which associate almost exclusively with neuronal cell bodies) to understand glia-soma interactions. We show that cortex glia tile with one another and with astrocytes to establish unique central nervous system (CNS) spatial domains that actively restrict glial growth, and selective ablation of cortex glia causes animal lethality. In an RNAi-based screen, we identified αSNAP (soluble NSF [N-ethylmalemeide-sensitive factor] attachment protein α) and several components of vesicle fusion and recycling machinery as essential for the maintenance of cortex glial morphology and continued contact with neurons. Interestingly, loss of the secreted neurotrophin Spätzle 3 (Spz3) phenocopied αSNAP phenotypes, which included loss of glial ensheathment of neuron cell bodies, increased neuronal cell death, and defects in animal behavior. Rescue experiments suggest that Spz3 can exert these effects only over very short distances. This work identifies essential roles for glial ensheathment of neuronal cell bodies in CNS homeostasis as well as Spz3 as a novel signaling factor required for maintenance of cortex glial morphology and neuron-glia contact.
Keywords: Drosophila; cortex glia; glia; neurotrophin; spz3; αSNAP.
© 2017 Coutinho-Budd et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Akagawa H, Hara Y, Togane Y, Iwabuchi K, Hiraoka T, Tsujimura H. 2015. The role of the effector caspases drICE and dcp-1 for cell death and corpse clearance in the developing optic lobe in Drosophila. Dev Biol 404: 61–75. - PubMed
-
- Akiyama-Oda Y, Hosoya T, Hotta Y. 1999. Asymmetric cell division of thoracic neuroblast 6-4 to bifurcate glial and neuronal lineage in Drosophila. Development 126: 1967–1974. - PubMed
-
- Allen NJ, Barres BA. 2009. Glia - more than just brain glue. Nature 457: 675–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
