A Conserved Cytoskeletal Signaling Cascade Mediates Neurotoxicity of FTDP-17 Tau Mutations In Vivo
- PMID: 29138281
- PMCID: PMC5761430
- DOI: 10.1523/JNEUROSCI.1550-17.2017
A Conserved Cytoskeletal Signaling Cascade Mediates Neurotoxicity of FTDP-17 Tau Mutations In Vivo
Abstract
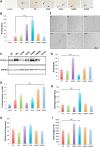
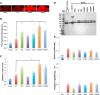
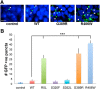
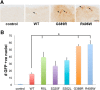
The microtubule binding protein tau is strongly implicated in multiple neurodegenerative disorders, including frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17), which is caused by mutations in tau. In vitro, FTDP-17 mutant versions of tau can reduce microtubule binding and increase the aggregation of tau, but the mechanism by which these mutations promote disease in vivo is not clear. Here we take a combined biochemical and in vivo modeling approach to define functional properties of tau driving neurotoxicity in vivo We express wild-type human tau and five FTDP-17 mutant forms of tau in Drosophila using a site-directed insertion strategy to ensure equivalent levels of expression. We then analyze multiple markers of neurodegeneration and neurotoxicity in transgenic animals, including analysis of both males and females. We find that FTDP-17 mutations act to enhance phosphorylation of tau and thus promote neurotoxicity in an in vivo setting. Further, we demonstrate that phosphorylation-dependent excess stabilization of the actin cytoskeleton is a key phosphorylation-dependent mediator of the toxicity of wild-type tau and of all the FTDP-17 mutants tested. Finally, we show that important downstream pathways, including autophagy and the unfolded protein response, are coregulated with neurotoxicity and actin cytoskeletal stabilization in brains of flies expressing wild-type human and various FTDP-17 tau mutants, supporting a conserved mechanism of neurotoxicity of wild-type tau and FTDP-17 mutant tau in disease pathogenesis.SIGNIFICANCE STATEMENT The microtubule protein tau aggregates and forms insoluble inclusion bodies known as neurofibrillary tangles in the brain tissue of patients with a variety of neurodegenerative disorders, including Alzheimer's disease. The tau protein is thus widely felt to play a key role in promoting neurodegeneration. However, precisely how tau becomes toxic is unclear. Here we capitalize on an "experiment of nature" in which rare missense mutations in tau cause familial neurodegeneration and neurofibrillary tangle formation. By comparing the biochemical activities of different tau mutations with their in vivo toxicity in a well controlled Drosophila model system, we find that all mutations tested increase phosphorylation of tau and trigger a cascade of neurotoxicity critically impinging on the integrity of the actin cytoskeleton.
Keywords: Alzheimer's; Drosophila; tau.
Copyright © 2018 the authors 0270-6474/18/380108-12$15.00/0.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
