First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin
- PMID: 29138298
- PMCID: PMC5686531
- DOI: 10.1128/mBio.00824-17
First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin
Abstract
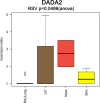
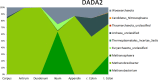
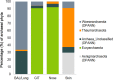
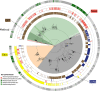
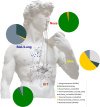
Human-associated archaea remain understudied in the field of microbiome research, although in particular methanogenic archaea were found to be regular commensals of the human gut, where they represent keystone species in metabolic processes. Knowledge on the abundance and diversity of human-associated archaea is extremely limited, and little is known about their function(s), their overall role in human health, or their association with parts of the human body other than the gastrointestinal tract and oral cavity. Currently, methodological issues impede the full assessment of the human archaeome, as bacteria-targeting protocols are unsuitable for characterization of the full spectrum of Archaea The goal of this study was to establish conservative protocols based on specifically archaea-targeting, PCR-based methods to retrieve first insights into the archaeomes of the human gastrointestinal tract, lung, nose, and skin. Detection of Archaea was highly dependent on primer selection and the sequence processing pipeline used. Our results enabled us to retrieve a novel picture of the human archaeome, as we found for the first time Methanobacterium and Woesearchaeota (DPANN superphylum) to be associated with the human gastrointestinal tract and the human lung, respectively. Similar to bacteria, human-associated archaeal communities were found to group biogeographically, forming (i) the thaumarchaeal skin landscape, (ii) the (methano)euryarchaeal gastrointestinal tract, (iii) a mixed skin-gastrointestinal tract landscape for the nose, and (iv) a woesearchaeal lung landscape. On the basis of the protocols we used, we were able to detect unexpectedly high diversity of archaea associated with different body parts.IMPORTANCE In summary, our study highlights the importance of the primers and data processing pipeline used to study the human archaeome. We were able to establish protocols that revealed the presence of previously undetected Archaea in all of the tissue samples investigated and to detect biogeographic patterns of the human archaeome in the gastrointestinal tract and on the skin and for the first time in the respiratory tract, i.e., the nose and lungs. Our results are a solid basis for further investigation of the human archaeome and, in the long term, discovery of the potential role of archaea in human health and disease.
Keywords: Archaea; archaeome; methanogens; microbiome.
Copyright © 2017 Koskinen et al.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
