Key Metabolites and Mechanistic Changes for Salt Tolerance in an Experimentally Evolved Sulfate-Reducing Bacterium, Desulfovibrio vulgaris
- PMID: 29138306
- PMCID: PMC5686539
- DOI: 10.1128/mBio.01780-17
Key Metabolites and Mechanistic Changes for Salt Tolerance in an Experimentally Evolved Sulfate-Reducing Bacterium, Desulfovibrio vulgaris
Abstract
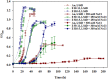
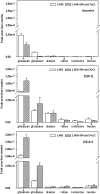
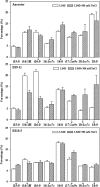
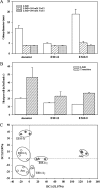
Rapid genetic and phenotypic adaptation of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough to salt stress was observed during experimental evolution. In order to identify key metabolites important for salt tolerance, a clone, ES10-5, which was isolated from population ES10 and allowed to experimentally evolve under salt stress for 5,000 generations, was analyzed and compared to clone ES9-11, which was isolated from population ES9 and had evolved under the same conditions for 1,200 generations. These two clones were chosen because they represented the best-adapted clones among six independently evolved populations. ES10-5 acquired new mutations in genes potentially involved in salt tolerance, in addition to the preexisting mutations and different mutations in the same genes as in ES9-11. Most basal abundance changes of metabolites and phospholipid fatty acids (PLFAs) were lower in ES10-5 than ES9-11, but an increase of glutamate and branched PLFA i17:1ω9c under high-salinity conditions was persistent. ES9-11 had decreased cell motility compared to the ancestor; in contrast, ES10-5 showed higher cell motility under both nonstress and high-salinity conditions. Both genotypes displayed better growth energy efficiencies than the ancestor under nonstress or high-salinity conditions. Consistently, ES10-5 did not display most of the basal transcriptional changes observed in ES9-11, but it showed increased expression of genes involved in glutamate biosynthesis, cation efflux, and energy metabolism under high salinity. These results demonstrated the role of glutamate as a key osmolyte and i17:1ω9c as the major PLFA for salt tolerance in D. vulgaris The mechanistic changes in evolved genotypes suggested that growth energy efficiency might be a key factor for selection.IMPORTANCE High salinity (e.g., elevated NaCl) is a stressor that affects many organisms. Salt tolerance, a complex trait involving multiple cellular pathways, is attractive for experimental evolutionary studies. Desulfovibrio vulgaris Hildenborough is a model sulfate-reducing bacterium (SRB) that is important in biogeochemical cycling of sulfur, carbon, and nitrogen, potentially for bio-corrosion, and for bioremediation of toxic heavy metals and radionuclides. The coexistence of SRB and high salinity in natural habitats and heavy metal-contaminated field sites laid the foundation for the study of salt adaptation of D. vulgaris Hildenborough with experimental evolution. Here, we analyzed a clone that evolved under salt stress for 5,000 generations and compared it to a clone evolved under the same condition for 1,200 generations. The results indicated the key roles of glutamate for osmoprotection and of i17:1ω9c for increasing membrane fluidity during salt adaptation. The findings provide valuable insights about the salt adaptation mechanism changes during long-term experimental evolution.
Keywords: Desulfovibrio vulgaris; PLFA; cell motility; energy efficiency; genomic mutations; organic solutes; transcriptomics.
Copyright © 2017 Zhou et al.
Figures
Similar articles
-
Rapid selective sweep of pre-existing polymorphisms and slow fixation of new mutations in experimental evolution of Desulfovibrio vulgaris.ISME J. 2015 Nov;9(11):2360-72. doi: 10.1038/ismej.2015.45. Epub 2015 Apr 7. ISME J. 2015. PMID: 25848870 Free PMC article.
-
Characterization of NaCl tolerance in Desulfovibrio vulgaris Hildenborough through experimental evolution.ISME J. 2013 Sep;7(9):1790-802. doi: 10.1038/ismej.2013.60. Epub 2013 Apr 11. ISME J. 2013. PMID: 23575373 Free PMC article.
-
Genetic Basis of Chromate Adaptation and the Role of the Pre-existing Genetic Divergence during an Experimental Evolution Study with Desulfovibrio vulgaris Populations.mSystems. 2021 Jun 29;6(3):e0049321. doi: 10.1128/mSystems.00493-21. Epub 2021 May 26. mSystems. 2021. PMID: 34061571 Free PMC article.
-
The adaptive genome of Desulfovibrio vulgaris Hildenborough.FEMS Microbiol Lett. 2006 Jul;260(2):127-33. doi: 10.1111/j.1574-6968.2006.00261.x. FEMS Microbiol Lett. 2006. PMID: 16842335 Review.
-
Maintenance of Cell Wall Integrity under High Salinity.Int J Mol Sci. 2021 Mar 23;22(6):3260. doi: 10.3390/ijms22063260. Int J Mol Sci. 2021. PMID: 33806816 Free PMC article. Review.
Cited by
-
Seven Years at High Salinity-Experimental Evolution of the Extremely Halotolerant Black Yeast Hortaea werneckii.J Fungi (Basel). 2021 Sep 4;7(9):723. doi: 10.3390/jof7090723. J Fungi (Basel). 2021. PMID: 34575761 Free PMC article.
-
Experimental evolution reveals nitrate tolerance mechanisms in Desulfovibrio vulgaris.ISME J. 2020 Nov;14(11):2862-2876. doi: 10.1038/s41396-020-00753-5. Epub 2020 Sep 15. ISME J. 2020. PMID: 32934357 Free PMC article.
-
Experimental Evolution as a High-Throughput Screen for Genetic Adaptations.mSphere. 2018 May 9;3(3):e00121-18. doi: 10.1128/mSphere.00121-18. eCollection 2018 May-Jun. mSphere. 2018. PMID: 29743200 Free PMC article.
-
OrpR is a σ54 -dependent activator using an iron-sulfur cluster for redox sensing in Desulfovibrio vulgaris Hildenborough.Mol Microbiol. 2021 Jul;116(1):231-244. doi: 10.1111/mmi.14705. Epub 2021 Feb 25. Mol Microbiol. 2021. PMID: 33595838 Free PMC article.
-
Bacterial community response to novel and repeated disturbances.Environ Microbiol Rep. 2024 Oct;16(5):e70022. doi: 10.1111/1758-2229.70022. Environ Microbiol Rep. 2024. PMID: 39387551 Free PMC article.
References
-
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BØ. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38:1406–1412. doi:10.1038/ng1906. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

