Phytochromes and Phytochrome Interacting Factors
- PMID: 29138351
- PMCID: PMC5813575
- DOI: 10.1104/pp.17.01384
Phytochromes and Phytochrome Interacting Factors
Abstract
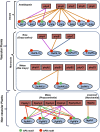
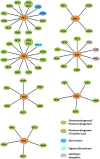
The basic helix-loop-helix domain-containing transcription factors that interact physically with the red and far-red light photoreceptors, phytochromes, are called PHYTOCHROME INTERACTING FACTORS (PIFs). In the last two decades, the phytochrome-PIF signaling module has been shown to be conserved from Physcomitrella patens to higher plants. Exciting recent studies highlight the discovery of at least four distinct kinases (PPKs, CK2, BIN2, and phytochrome itself) and four families of ubiquitin ligases (SCFEBF1/2, CUL3LRB, CUL3BOP, and CUL4COP1-SPA) that regulate PIF abundance both in dark and light conditions. This review discusses these recent discoveries with a focus on the central phytochrome signaling mechanisms that have a profound impact on plant growth and development in response to light.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 - PubMed
-
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 - PubMed
-
- Bu Q, Castillon A, Chen F, Zhu L, Huq E (2011a) Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol Biol 77: 501–511 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

