Timescale Separation of Positive and Negative Signaling Creates History-Dependent Responses to IgE Receptor Stimulation
- PMID: 29138425
- PMCID: PMC5686181
- DOI: 10.1038/s41598-017-15568-2
Timescale Separation of Positive and Negative Signaling Creates History-Dependent Responses to IgE Receptor Stimulation
Abstract
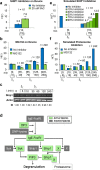
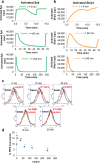
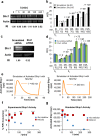
The high-affinity receptor for IgE expressed on the surface of mast cells and basophils interacts with antigens, via bound IgE antibody, and triggers secretion of inflammatory mediators that contribute to allergic reactions. To understand how past inputs (memory) influence future inflammatory responses in mast cells, a microfluidic device was used to precisely control exposure of cells to alternating stimulatory and non-stimulatory inputs. We determined that the response to subsequent stimulation depends on the interval of signaling quiescence. For shorter intervals of signaling quiescence, the second response is blunted relative to the first response, whereas longer intervals of quiescence induce an enhanced second response. Through an iterative process of computational modeling and experimental tests, we found that these memory-like phenomena arise from a confluence of rapid, short-lived positive signals driven by the protein tyrosine kinase Syk; slow, long-lived negative signals driven by the lipid phosphatase Ship1; and slower degradation of Ship1 co-factors. This work advances our understanding of mast cell signaling and represents a generalizable approach for investigating the dynamics of signaling systems.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
SHIP1 and the negative control of mast cell/basophil activation by supra-optimal antigen concentrations.Mol Immunol. 2015 Jan;63(1):32-7. doi: 10.1016/j.molimm.2014.02.017. Epub 2014 Mar 25. Mol Immunol. 2015. PMID: 24679713 Review.
-
Trans-inhibition of activation and proliferation signals by Fc receptors in mast cells and basophils.Sci Signal. 2016 Dec 20;9(459):ra126. doi: 10.1126/scisignal.aag1401. Sci Signal. 2016. PMID: 27999175
-
Immunologically mediated signaling in basophils and mast cells: finding therapeutic targets for allergic diseases in the human FcvarepsilonR1 signaling pathway.Immunopharmacology. 2000 Jul 25;48(3):269-81. doi: 10.1016/s0162-3109(00)00224-1. Immunopharmacology. 2000. PMID: 10960668 Review.
-
Antigen-induced reduction in mast cell and basophil functional responses due to reduced Syk protein levels.Int Arch Allergy Immunol. 2005 Sep;138(1):29-39. doi: 10.1159/000087355. Epub 2005 Aug 8. Int Arch Allergy Immunol. 2005. PMID: 16088210
-
FcepsilonRI-alpha siRNA inhibits the antigen-induced activation of mast cells.Iran J Allergy Asthma Immunol. 2009 Dec;8(4):177-83. Iran J Allergy Asthma Immunol. 2009. PMID: 20404387
Cited by
-
Computational Methods for the Design of Recombinase Logic Circuits with Adaptable Circuit Specifications.Methods Mol Biol. 2023;2553:155-171. doi: 10.1007/978-1-0716-2617-7_8. Methods Mol Biol. 2023. PMID: 36227543
-
The Constrained Disorder Principle Overcomes the Challenges of Methods for Assessing Uncertainty in Biological Systems.J Pers Med. 2024 Dec 28;15(1):10. doi: 10.3390/jpm15010010. J Pers Med. 2024. PMID: 39852203 Free PMC article. Review.
-
Erroneous detection of desensitization doses in the prevention of hypersensitivity reactions.BMC Res Notes. 2023 Feb 3;16(1):12. doi: 10.1186/s13104-023-06278-2. BMC Res Notes. 2023. PMID: 36737795 Free PMC article.
-
Bayesian inference using qualitative observations of underlying continuous variables.Bioinformatics. 2020 May 1;36(10):3177-3184. doi: 10.1093/bioinformatics/btaa084. Bioinformatics. 2020. PMID: 32049328 Free PMC article.
-
New Mechanistic Advances in FcεRI-Mast Cell-Mediated Allergic Signaling.Clin Rev Allergy Immunol. 2022 Dec;63(3):431-446. doi: 10.1007/s12016-022-08955-9. Epub 2022 Oct 17. Clin Rev Allergy Immunol. 2022. PMID: 36251242 Free PMC article. Review.
References
-
- Weetall M, Holowka D, Baird B. Heterologous desensitization of the high affinity receptor for IgE (Fc epsilon R1) on RBL cells. Journal of immunology (Baltimore, Md. : 1950) 1993;150:4072–4083. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

