The functions and unique features of long intergenic non-coding RNA
- PMID: 29138516
- PMCID: PMC5889127
- DOI: 10.1038/nrm.2017.104
The functions and unique features of long intergenic non-coding RNA
Abstract
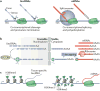
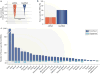
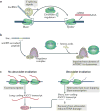
Long intergenic non-coding RNA (lincRNA) genes have diverse features that distinguish them from mRNA-encoding genes and exercise functions such as remodelling chromatin and genome architecture, RNA stabilization and transcription regulation, including enhancer-associated activity. Some genes currently annotated as encoding lincRNAs include small open reading frames (smORFs) and encode functional peptides and thus may be more properly classified as coding RNAs. lincRNAs may broadly serve to fine-tune the expression of neighbouring genes with remarkable tissue specificity through a diversity of mechanisms, highlighting our rapidly evolving understanding of the non-coding genome.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Venter CJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. - PubMed
-
- Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

