Visualizing the GPCR Network: Classification and Evolution
- PMID: 29138525
- PMCID: PMC5686146
- DOI: 10.1038/s41598-017-15707-9
Visualizing the GPCR Network: Classification and Evolution
Abstract
In this study, we delineate an unsupervised clustering algorithm, minimum span clustering (MSC), and apply it to detect G-protein coupled receptor (GPCR) sequences and to study the GPCR network using a base dataset of 2770 GPCR and 652 non-GPCR sequences. High detection accuracy can be achieved with a proper dataset. The clustering results of GPCRs derived from MSC show a strong correlation between their sequences and functions. By comparing our level 1 MSC results with the GPCRdb classification, the consistency is 87.9% for the fourth level of GPCRdb, 89.2% for the third level, 98.4% for the second level, and 100% for the top level (the lowest resolution level of GPCRdb). The MSC results of GPCRs can be well explained by estimating the selective pressure of GPCRs, as exemplified by investigating the largest two subfamilies, peptide receptors (PRs) and olfactory receptors (ORs), in class A GPCRs. PRs are decomposed into three groups due to a positive selective pressure, whilst ORs remain as a single group due to a negative selective pressure. Finally, we construct and compare phylogenetic trees using distance-based and character-based methods, a combination of which could convey more comprehensive information about the evolution of GPCRs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
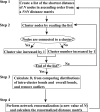
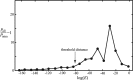





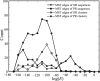


References
-
- Schonenbach NS, Hussain S, O’Malley MA. Structure and function of G protein-coupled receptor oligomers: implications for drug discovery. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2015;7:408–427. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

