miR‑185 inhibits non‑small cell lung cancer cell proliferation and invasion through targeting of SOX9 and regulation of Wnt signaling
- PMID: 29138830
- PMCID: PMC5780119
- DOI: 10.3892/mmr.2017.8050
miR‑185 inhibits non‑small cell lung cancer cell proliferation and invasion through targeting of SOX9 and regulation of Wnt signaling
Erratum in
-
[Corrigedum] miR‑185 inhibits non‑small cell lung cancer cell proliferation and invasion through targeting of SOX9 and regulation of Wnt signaling.Mol Med Rep. 2023 May;27(5):97. doi: 10.3892/mmr.2023.12984. Epub 2023 Mar 24. Mol Med Rep. 2023. PMID: 36960863 Free PMC article.
Abstract
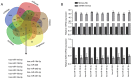
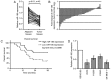
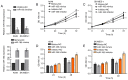
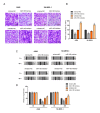
SRY-box 9 (SOX9) is an important transcription factor required for development, which has additionally been reported to be an independent prognostic indicator for the survival of patients with non‑small cell lung cancer (NSCLC). Accumulating evidence has indicated that dysregulation of microRNAs (miRNAs/miRs) may contribute to the initiation and progression of cancer. SOX9 may be regulated by a number of miRNAs in different types of cancer, including in NSCLC. The present study sought to identify novel candidate miRNAs associated with SOX9 in NSCLC using online tools, and investigated the detailed functions of miR‑185, which suppressed SOX9 mRNA expression most strongly out of the candidate miRNAs. It was observed that ectopic miR‑185 expression significantly suppressed NSCLC cell proliferation, invasion and migration. Using luciferase reporter gene and RNA immunoprecipitation assays, SOX9 was confirmed to be a direct target of miR‑185. In addition, the downstream Wnt signaling‑associated factors β‑catenin and c‑Myc proto‑oncogene protein (Myc) were demonstrated to be inhibited by miR‑185 overexpression. SOX9, β‑catenin and c‑Myc mRNA expression was significantly upregulated in NSCLC tissues, and was inversely correlated with miR‑185 expression. The results of the present study demonstrated that rescuing miR‑185 expression in NSCLC, thereby inhibiting SOX9 expression and the downstream Wnt signaling, and leading to the suppression of NSCLC cell proliferation, invasion and migration, may be a promising strategy for the treatment of NSCLC.
Figures

Similar articles
-
Long non-coding RNA PVT1 contributes to cell growth and metastasis in non-small-cell lung cancer by regulating miR-361-3p/SOX9 axis and activating Wnt/β-catenin signaling pathway.Biomed Pharmacother. 2020 Jun;126:110100. doi: 10.1016/j.biopha.2020.110100. Epub 2020 Mar 24. Biomed Pharmacother. 2020. PMID: 32197208
-
Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway.Oncotarget. 2017 Mar 14;8(11):17785-17794. doi: 10.18632/oncotarget.14854. Oncotarget. 2017. PMID: 28147312 Free PMC article.
-
Overexpression of HIF-2α-Dependent NEAT1 Promotes the Progression of Non-Small Cell Lung Cancer through miR-101-3p/SOX9/Wnt/β-Catenin Signal Pathway.Cell Physiol Biochem. 2019;52(3):368-381. doi: 10.33594/000000026. Epub 2019 Mar 8. Cell Physiol Biochem. 2019. PMID: 30845377
-
SOX9: The master regulator of cell fate in breast cancer.Biochem Pharmacol. 2020 Apr;174:113789. doi: 10.1016/j.bcp.2019.113789. Epub 2020 Jan 3. Biochem Pharmacol. 2020. PMID: 31911091 Free PMC article. Review.
-
Role of miRNAs in lung cancer.J Cell Physiol. 2018 Apr 20. doi: 10.1002/jcp.26607. Online ahead of print. J Cell Physiol. 2018. Retraction in: J Cell Physiol. 2025 Jan;240(1):e31519. doi: 10.1002/jcp.31519. PMID: 29676470 Retracted. Review.
Cited by
-
lncRNA RPPH1 promotes non-small cell lung cancer progression through the miR-326/WNT2B axis.Oncol Lett. 2020 Oct;20(4):105. doi: 10.3892/ol.2020.11966. Epub 2020 Aug 10. Oncol Lett. 2020. PMID: 32831924 Free PMC article.
-
MicroRNA Signatures Associated with Bronchopulmonary Dysplasia Severity in Tracheal Aspirates of Preterm Infants.Biomedicines. 2021 Mar 5;9(3):257. doi: 10.3390/biomedicines9030257. Biomedicines. 2021. PMID: 33807742 Free PMC article.
-
Porcupine inhibitor LGK‑974 inhibits Wnt/β‑catenin signaling and modifies tumor‑associated macrophages resulting in inhibition of the malignant behaviors of non‑small cell lung cancer cells.Mol Med Rep. 2021 Aug;24(2):550. doi: 10.3892/mmr.2021.12189. Epub 2021 Jun 3. Mol Med Rep. 2021. PMID: 34080032 Free PMC article.
-
Behavioral Changes in Stem-Cell Potency by HepG2-Exhausted Medium.Cells. 2020 Aug 12;9(8):1890. doi: 10.3390/cells9081890. Cells. 2020. PMID: 32806709 Free PMC article.
-
RIF1 promotes tumor growth and cancer stem cell-like traits in NSCLC by protein phosphatase 1-mediated activation of Wnt/β-catenin signaling.Cell Death Dis. 2018 Sep 20;9(10):942. doi: 10.1038/s41419-018-0972-4. Cell Death Dis. 2018. PMID: 30237512 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

