Genomic analysis of atypical fibroxanthoma
- PMID: 29141020
- PMCID: PMC5687749
- DOI: 10.1371/journal.pone.0188272
Genomic analysis of atypical fibroxanthoma
Abstract
Atypical fibroxanthoma (AFX), is a rare type of skin cancer affecting older individuals with sun damaged skin. Since there is limited genomic information about AFX, our study seeks to improve the understanding of AFX through whole-exome and RNA sequencing of 8 matched tumor-normal samples. AFX is a highly mutated malignancy with recurrent mutations in a number of genes, including COL11A1, ERBB4, CSMD3, and FAT1. The majority of mutations identified were UV signature (C>T in dipyrimidines). We observed deletion of chromosomal segments on chr9p and chr13q, including tumor suppressor genes such as KANK1 and CDKN2A, but no gene fusions were found. Gene expression profiling revealed several biological pathways that are upregulated in AFX, including tumor associated macrophage response, GPCR signaling, and epithelial to mesenchymal transition (EMT). To further investigate the presence of EMT in AFX, we conducted a gene expression meta-analysis that incorporated RNA-seq data from dermal fibroblasts and keratinocytes. Ours is the first study to employ high throughput sequencing for molecular profiling of AFX. These data provide valuable insights to inform models of carcinogenesis and additional research towards tumor-directed therapy.
Conflict of interest statement
Figures

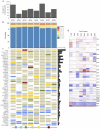

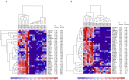

References
-
- Anderson HL, Joseph AK. A pilot feasibility study of a rare skin tumor database. Dermatol Surg Off Publ Am Soc Dermatol Surg Al. 2007;33: 693–696. doi: 10.1111/j.1524-4725.2007.33145.x - DOI - PubMed
-
- Wieland CN, Dyck R, Weenig RH, Comfere NI. The role of CD10 in distinguishing atypical fibroxanthoma from sarcomatoid (spindle cell) squamous cell carcinoma. J Cutan Pathol. 2011;38: 884–888. doi: 10.1111/j.1600-0560.2011.01768.x - DOI - PubMed
-
- Ziemer M. Atypical fibroxanthoma. JDDG J Dtsch Dermatol Ges. 2012;10: 537–548. doi: 10.1111/j.1610-0387.2012.07980.x - DOI - PubMed
-
- Simon MP, Pedeutour F, Sirvent N, Grosgeorge J, Minoletti F, Coindre JM, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15: 95–98. doi: 10.1038/ng0197-95 - DOI - PubMed
-
- Mirza B, Weedon D. Atypical fibroxanthoma: a clinicopathological study of 89 cases. Australas J Dermatol. 2005;46: 235–238. doi: 10.1111/j.1440-0960.2005.00190.x - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

