Rescue of high-specificity Cas9 variants using sgRNAs with matched 5' nucleotides
- PMID: 29141659
- PMCID: PMC5686910
- DOI: 10.1186/s13059-017-1355-3
Rescue of high-specificity Cas9 variants using sgRNAs with matched 5' nucleotides
Abstract
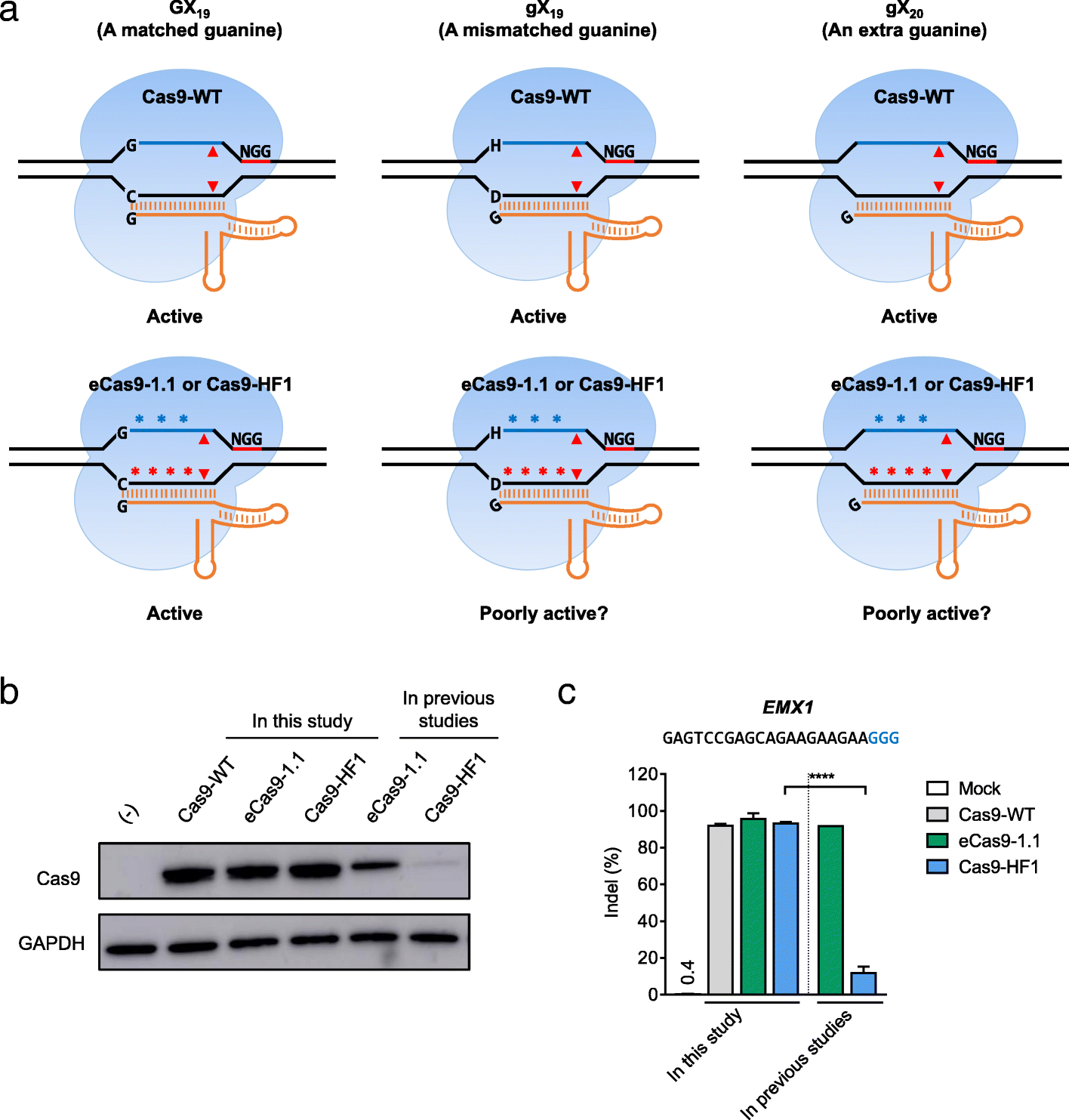
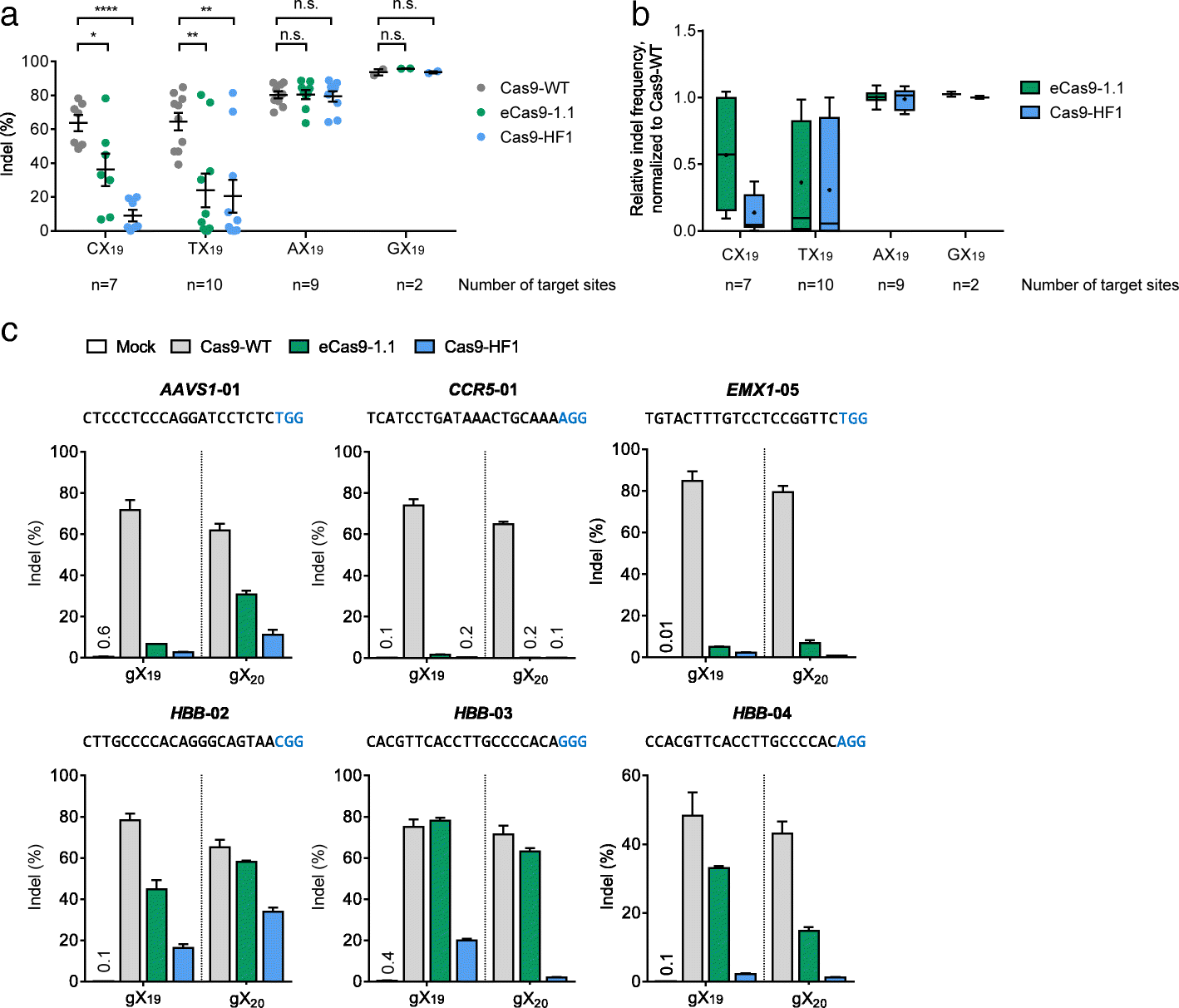
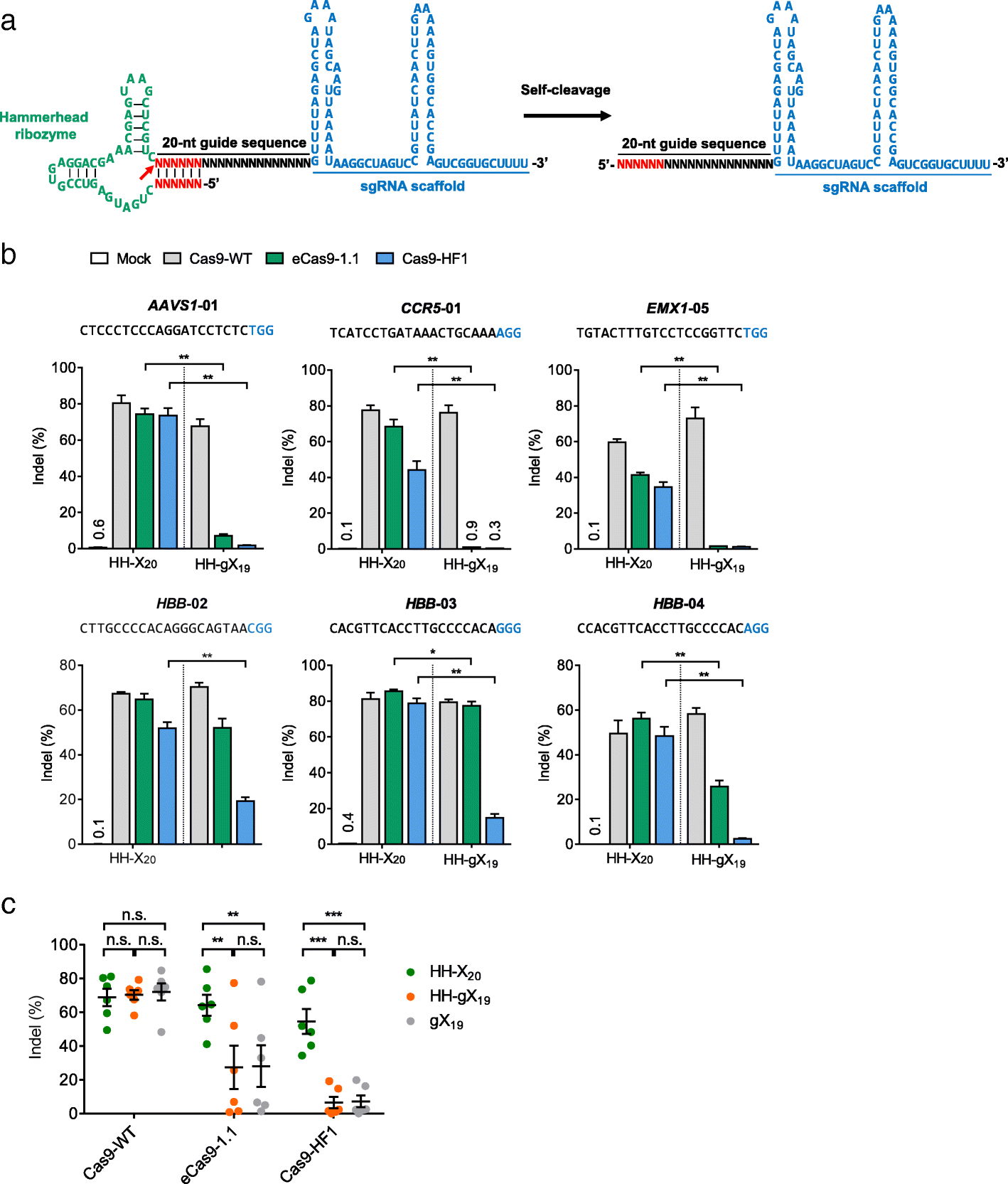
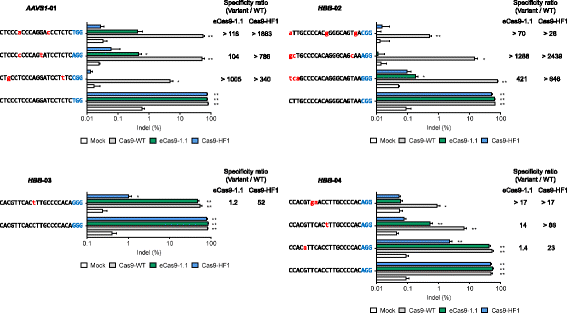
We report that engineered Cas9 variants with improved specificity-eCas9-1.1 and Cas9-HF1-are often poorly active in human cells, when complexed with single guide RNAs (sgRNAs) with a mismatch at the 5' terminus, relative to target DNA sequences. Because the nucleotide at the 5' end of sgRNAs, expressed under the control of the commonly-used U6 promoter, is fixed to a guanine, these attenuated Cas9 variants are not useful at many target sites. By using sgRNAs with matched 5' nucleotides, produced by linking them to a self-cleaving ribozyme, the editing activity of Cas9 variants can be rescued without sacrificing high specificity.
Keywords: CRISPR-Cas; Engineered Cas9 variants; Hammerhead ribozyme-linked sgRNA; Off-target effect.
Conflict of interest statement
Ethics approval and consent to participate
No ethics approval was required for this study.
Competing interests
J-SK is a co-founder of and holds stocks in ToolGen, Inc.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

