DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development
- PMID: 29141958
- PMCID: PMC6839835
- DOI: 10.1136/gutjnl-2017-314817
DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development
Abstract
Objective: Human intestinal epithelial organoids (IEOs) are increasingly being recognised as a highly promising translational research tool. However, our understanding of their epigenetic molecular characteristics and behaviour in culture remains limited.
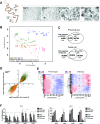
Design: We performed genome-wide DNA methylation and transcriptomic profiling of human IEOs derived from paediatric/adult and fetal small and large bowel as well as matching purified human gut epithelium. Furthermore, organoids were subjected to in vitro differentiation and genome editing using CRISPR/Cas9 technology.
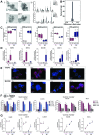
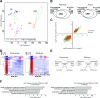
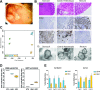
Results: We discovered stable epigenetic signatures which define regional differences in gut epithelial function, including induction of segment-specific genes during cellular differentiation. Established DNA methylation profiles were independent of cellular environment since organoids retained their regional DNA methylation over prolonged culture periods. In contrast to paediatric and adult organoids, fetal gut-derived organoids showed distinct dynamic changes of DNA methylation and gene expression in culture, indicative of an in vitro maturation. By applying CRISPR/Cas9 genome editing to fetal organoids, we demonstrate that this process is partly regulated by TET1, an enzyme involved in the DNA demethylation process. Lastly, generating IEOs from a child diagnosed with gastric heterotopia revealed persistent and distinct disease-associated DNA methylation differences, highlighting the use of organoids as disease-specific research models.
Conclusions: Our study demonstrates striking similarities of epigenetic signatures in mucosa-derived IEOs with matching primary epithelium. Moreover, these results suggest that intestinal stem cell-intrinsic DNA methylation patterns establish and maintain regional gut specification and are involved in early epithelial development and disease.
Keywords: intestinal cell lines; intestinal development; intestinal epithelium; intestinal gene regulation; intestinal stem cell.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2019. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures



Comment in
-
Developmental biology: Stable epigenetic signatures in intestinal organoids.Nat Rev Gastroenterol Hepatol. 2018 Jan;15(1):4. doi: 10.1038/nrgastro.2017.179. Epub 2017 Dec 13. Nat Rev Gastroenterol Hepatol. 2018. PMID: 29235548 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
