ArfGAP1 restricts Mycobacterium tuberculosis entry by controlling the actin cytoskeleton
- PMID: 29141986
- PMCID: PMC5757213
- DOI: 10.15252/embr.201744371
ArfGAP1 restricts Mycobacterium tuberculosis entry by controlling the actin cytoskeleton
Abstract
The interaction of Mycobacterium tuberculosis (Mtb) with pulmonary epithelial cells is critical for early stages of bacillus colonization and during the progression of tuberculosis. Entry of Mtb into epithelial cells has been shown to depend on F-actin polymerization, though the molecular mechanisms are still unclear. Here, we demonstrate that mycobacterial uptake into epithelial cells requires rearrangements of the actin cytoskeleton, which are regulated by ADP-ribosylation factor 1 (Arf1) and phospholipase D1 (PLD1), and is dependent on the M3 muscarinic receptor (M3R). We show that this pathway is controlled by Arf GTPase-activating protein 1 (ArfGAP1), as its silencing has an impact on actin cytoskeleton reorganization leading to uncontrolled uptake and replication of Mtb. Furthermore, we provide evidence that this pathway is critical for mycobacterial entry, while the cellular infection with other pathogens, such as Shigella flexneri and Yersinia pseudotuberculosis, is not affected. Altogether, these results reveal how cortical actin plays the role of a barrier to prevent mycobacterial entry into epithelial cells and indicate a novel role for ArfGAP1 as a restriction factor of host-pathogen interactions.
Keywords: Mycobacterium tuberculosis; Arf1; ArfGAP1; epithelial cell; phospholipase D1.
© 2017 The Authors.
Figures

- A, B
A549 epithelial cells were transfected with pooled siRNA in 384‐well plates and infected with Mtb H37Rv‐GFP at an MOI of 5. Five days post‐infection, images were acquired by automated confocal microscopy followed by image analysis. Shown are the applied workflow (A) and a Z score scatter plot of the obtained data (B, upper panel). Among the most significant siRNA hits, ArfGAP1 was identified in Mtb‐infected cells. A representative confocal image of scramble and siArfGAP1‐transfected cells is shown below (B, lower panel). Cell nuclei were stained by DAPI (blue), while GFP‐expressing Mtb are shown in green. Scale bars: 50 μm.
- C, D
The impact of ArfGAP1 silencing on Mtb uptake (C) and intracellular replication of Mtb (D) were analyzed in infected A549 cells and by automated confocal microscopy. Shown are the applied workflow (upper panel) and results of three independent experiments (lower panel). Both overall infection rates (left histogram in C and D) and bacterial load per cell (right histogram in C and D) are indicated. Data analysis was carried out to obtain percentages of infected cells (n ≥ 700) and quantified areas of intracellular bacteria (px: pixels). Data are presented as mean ± SEM. ***P < 0.0005, ns: not significant (Student's t‐test).

- A, B
Scramble and ArfGAP1 KD cells were infected with Mtb H37Ra‐GFP at MOI 10 and analyzed 4 days post‐infection by automated confocal microscopy. Shown are representative images (A) and Mtb colonization of cells (B). Cell nuclei were stained by DAPI (blue), while GFP‐expressing Mtb are shown in green. Scale bars: 50 μm.
- C, D
Similarly, these cells were also lysed and plated at different dilutions (C) to determine colony‐forming units (CFU) (D).
- E
The impact of ArfGAP1 silencing on Mtb H37Ra uptake at MOI 5 was analyzed by automated confocal microscopy at the indicated time points post‐infection.
- F
A549 cells were treated with the ArfGAP1 inhibitor QS11 and analyzed for Mtb H37Ra colonization at MOI 5 after 4 h post‐infection.
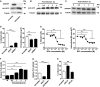
- A–C
A549 cells were transfected with scramble and ArfGAP1 siRNA and analyzed for active Arf1‐GTP by pull‐down and Western blotting (A). Tubulin was used as loading control. Similarly, Arf1‐GTP was analyzed in cells infected with Mtb H37Rv at an MOI of 5 at the indicated times post‐infection (B). Mtb‐infected cells were also analyzed for the expression of ArfGAP1 by Western blotting (C). Shown are representative blots of two independent experiments.
- D
A549 cells were transfected with scramble, Arf1, and GRP1 siRNA, and Mtb colonization at MOI 20 was analyzed after 4 h.
- E
A549 cells were treated with the indicated concentrations of BFA for 18 h prior infection at MOI 20, and Mtb colonization was analyzed after 4 h by automated confocal microscopy.
- F, G
PLD activity was measured enzymatically using the Amplex Red reagent in Mtb‐infected A549 cells at MOI 5 (F) and in cells that were transfected with scramble, Arf1, and ArfGAP1 siRNA (G) (n = 3).
- H
Mtb colonization at MOI 20 was also analyzed after 4 h by automated confocal microscopy in cells transfected with scramble, PLD1‐specific siRNA, and PLD2‐specific siRNA.

Known relationships of Arf1 with different receptors obtained by Ingenuity Pathway Analysis (IPA).
ArfGAP1 KD cells were transfected with scramble siRNA and the indicated targeting siRNA and analyzed for Mtb H37Ra colonization at MOI 20 4 h post‐infection. Data are presented as mean ± SEM. ***P < 0.0005, *P < 0.05, ns: not significant (Student's t‐test). Histograms display at least 400 analyzed cells in six replicates. Shown are representative examples of two independent experiments.

- A, B
Impact of M3R silencing on Mtb H37Rv colonization in A549 cells at MOI 20 was analyzed after 4 h by automated confocal microscopy. Shown are representative images (A), overall infection rates (left histogram in B), and bacterial load per cell (right histogram in B) in the absence and presence of additional ArfGAP1 silencing. Scale bar: 25 μm.
- C
Mtb infection at MOI 20 was also analyzed after 4 h in ArfGAP1 KD cells using the M3R inhibitor Zamifenacin.
- D
Active Arf1‐GTP was measured by pull‐down and Western blotting in A549 cells transfected with scramble and M3R siRNA. Shown are a representative WB image (left panel) and the quantification of two independent experiments (right panel).
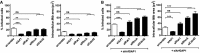
- A, B
A549 cells were transfected with scramble siRNA and the indicated targeting siRNA in the absence (A) and presence of additional ArfGAP1 silencing (B). Mtb H37Rv colonization of transfected cells at MOI 20 was analyzed by automated confocal microscopy 4 h post‐infection. Data are presented as mean ± SEM. ***P < 0.0005, **P < 0.005, ns: not significant (Student's t‐test). Histograms display at least 400 analyzed cells in six replicates. Shown are representative examples of two independent experiments.

- A, B
Scramble and ArfGAP1 KD cells were transfected with GFP‐actin, and images were recorded by automated confocal microscopy at the indicated time points during Mtb H37Rv infection at MOI 5 (A). Insets of detailed areas (white frames) are shown on the right. Scale bar: 20 μm. The average number of actin patches per cell was quantified in scramble siRNA cells over time (n = 7) (B).
- C
Cellular F‐actin distribution of scramble, ArfGAP1 KD, Arf1 KD, and M3R KD cells was analyzed by SIM microscopy. F‐actin was labeled by phalloidin, and nuclei were stained by DAPI. Scale bars: 5 μm.
- D, E
Phalloidin‐labeled cells were also analyzed by automated confocal microscopy (n ≥ 1,000) and quantified for the presence of cortical F‐actin (D) and actin stress fibers (E). Shown are representative examples of two independent experiments.
- F
Cellular elasticity of scramble and ArfGAP1 KD cells was measured by atomic force microscopy. Histograms display 42 analyzed cells. Shown are representative examples of three independent experiments.
- G
Scramble and ArfGAP1 KD cells were also treated with 0.8 μM cytochalasin D (CytoD) and Mtb colonization at MOI 20 was analyzed after 4 h by automated confocal microscopy.

- A
Scramble and ArfGAP1 KD epithelial cells (HeLa and A549), differentiated monocytes (THP‐1), and primary macrophages (HuMac) were analyzed by Western blotting for the expression of ArfGAP1. β‐actin and tubulin were used as loading control.
- B
siRNA‐transfected A549 cells were infected with Mtb H37Ra and analyzed by automated confocal microscopy. siRNA‐transfected HeLa and A549 cells were infected at an MOI of 20, while THP‐1 and HuMac were infected at MOI 2. Mtb colonization was analyzed after 4 h by automated microscopy.
- C, D
Transfected HeLa cells were analyzed for colonization by Shigella flexneri (C) and Yersinia pseudotuberculosis (D) at two different MOI.
- E
Proposed working model of actin rearrangements and Mtb entry in the presence (left panel) and absence of ArfGAP1 (right panel). See text for further details.

- A–C
Scramble and ArfGAP1 KD HeLa cells were labeled by phalloidin and analyzed by automated confocal microscopy. Shown are representative images (A) and cells (n ≥ 700) quantified for the presence of cortical F‐actin (B) and actin stress fibers (C).
- D, E
Actin expression was measured by Western blotting in A549 cells transfected with scramble and ArfGAP1 siRNA. Tubulin was used as loading control. Shown are one representative WB image (D) and the quantification of actin expression normalized to the expression of tubulin of three independent experiments (E).

- A, B
Mycobacterial colonization of scramble and ArfGAP1 KD A549 cells infected with Mycobacterium bovis BCG (A) and M. bovis BCG ∆hbhA (B) at MOI 1 4 h post‐infection analyzed by automated confocal microscopy. Data are presented as mean ± SEM. ***P < 0.0005, **P < 0.005 (Student's t‐test). Histograms display at least 400 analyzed cells in six replicates. Shown are representative examples of two independent experiments.
References
-
- Cossart P, Sansonetti PJ (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304: 242–248 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

