Interaction between a Unique Minor Protein and a Major Capsid Protein of Bluetongue Virus Controls Virus Infectivity
- PMID: 29142128
- PMCID: PMC5774872
- DOI: 10.1128/JVI.01784-17
Interaction between a Unique Minor Protein and a Major Capsid Protein of Bluetongue Virus Controls Virus Infectivity
Abstract
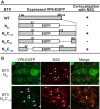
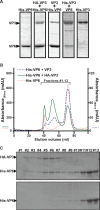
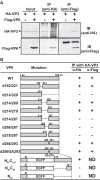
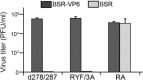
Among the Reoviridae family of double-stranded RNA viruses, only members of the Orbivirus genus possess a unique structural protein, termed VP6, within their particles. Bluetongue virus (BTV), an important livestock pathogen, is the prototype Orbivirus BTV VP6 is an ATP-dependent RNA helicase, and it is indispensable for virus replication. In the study described in this report, we investigated how VP6 might be recruited to the virus capsid and whether the BTV structural protein VP3, which forms the internal layer of the virus capsid core, is involved in VP6 recruitment. We first demonstrated that VP6 interacts with VP3 and colocalizes with VP3 during capsid assembly. A series of VP6 mutants was then generated, and in combination with immunoprecipitation and size exclusion chromatographic analyses, we demonstrated that VP6 directly interacts with VP3 via a specific region of the C-terminal portion of VP6. Finally, using our reverse genetics system, mutant VP6 proteins were introduced into the BTV genome and interactions between VP6 and VP3 were shown in a live cell system. We demonstrate that BTV strains possessing a mutant VP6 are replication deficient in wild-type BSR cells and fail to recruit the viral replicase complex into the virus particle core. Taken together, these data suggest that the interaction between VP3 and VP6 could be important in the packaging of the viral genome and early stages of particle formation.IMPORTANCE The orbivirus bluetongue virus (BTV) is the causative agent of bluetongue disease of livestock, often causing significant economic and agricultural impacts in the livestock industry. In the study described in this report, we identified the essential region and residues of the unique orbivirus capsid protein VP6 which are responsible for its interaction with other BTV proteins and its subsequent recruitment into the virus particle. The nature and mechanism of these interactions suggest that VP6 has a key role in packaging of the BTV genome into the virus particle. As such, this is a highly significant finding, as this new understanding of BTV assembly could be exploited to design novel vaccines and antivirals against bluetongue disease.
Keywords: BTV; VP6 translocation; reverse genetics system.
Copyright © 2018 Matsuo et al.
Figures


References
-
- Roy P. 2013. Orbiviruses, p 1402–1423. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Matsuo E, Celma CC, Boyce M, Viarouge C, Sailleau C, Dubois E, Breard E, Thiery R, Zientara S, Roy P. 2011. Generation of replication-defective virus-based vaccines that confer full protection in sheep against virulent bluetongue virus challenge. J Virol 85:10213–10221. doi:10.1128/JVI.05412-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

