Transient cardiomyocyte fusion regulates cardiac development in zebrafish
- PMID: 29142194
- PMCID: PMC5688123
- DOI: 10.1038/s41467-017-01555-8
Transient cardiomyocyte fusion regulates cardiac development in zebrafish
Abstract
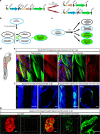
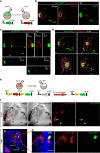
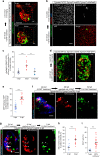
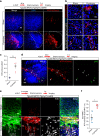
Cells can sacrifice their individuality by fusing, but the prevalence and significance of this process are poorly understood. To approach these questions, here we generate transgenic reporter lines in zebrafish to label and specifically ablate fused cells. In addition to skeletal muscle cells, the reporters label cardiomyocytes starting at an early developmental stage. Genetic mosaics generated by cell transplantation show cardiomyocytes expressing both donor- and host-derived transgenes, confirming the occurrence of fusion in larval hearts. These fusion events are transient and do not generate multinucleated cardiomyocytes. Functionally, cardiomyocyte fusion correlates with their mitotic activity during development as well as during regeneration in adult animals. By analyzing the cell fusion-compromised jam3b mutants, we propose a role for membrane fusion in cardiomyocyte proliferation and cardiac function. Together, our findings uncover the previously unrecognized process of transient cardiomyocyte fusion and identify its potential role in cardiac development and function.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

