Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma
- PMID: 29142209
- PMCID: PMC5688124
- DOI: 10.1038/s41467-017-01199-8
Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma
Abstract
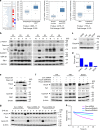
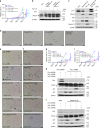
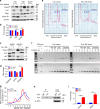
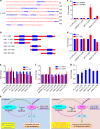
The Fbxo4 tumour suppressor is a component of an Skp1-Cul1-F-box E3 ligase for which two substrates are known. Here we show purification of SCFFbxo4 complexes results in the identification of fragile X protein family (FMRP, Fxr1 and Fxr2) as binding partners. Biochemical and functional analyses reveal that Fxr1 is a direct substrate of SCFFbxo4. Consistent with a substrate relationship, Fxr1 is overexpressed in Fbxo4 knockout cells, tissues and in human cancer cells, harbouring inactivating Fbxo4 mutations. Critically, in head and neck squamous cell carcinoma, Fxr1 overexpression correlates with reduced Fbxo4 levels in the absence of mutations or loss of mRNA, suggesting the potential for feedback regulation. Direct analysis reveals that Fbxo4 translation is attenuated by Fxr1, indicating the existence of a feedback loop that contributes to Fxr1 overexpression and the loss of Fbxo4. Ultimately, the consequence of Fxr1 overexpression is the bypass of senescence and neoplastic progression.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

