Unique transcriptome signatures and GM-CSF expression in lymphocytes from patients with spondyloarthritis
- PMID: 29142230
- PMCID: PMC5688161
- DOI: 10.1038/s41467-017-01771-2
Unique transcriptome signatures and GM-CSF expression in lymphocytes from patients with spondyloarthritis
Abstract
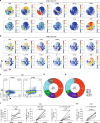
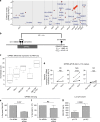
Spondyloarthritis encompasses a group of common inflammatory diseases thought to be driven by IL-17A-secreting type-17 lymphocytes. Here we show increased numbers of GM-CSF-producing CD4 and CD8 lymphocytes in the blood and joints of patients with spondyloarthritis, and increased numbers of IL-17A+GM-CSF+ double-producing CD4, CD8, γδ and NK cells. GM-CSF production in CD4 T cells occurs both independently and in combination with classical Th1 and Th17 cytokines. Type 3 innate lymphoid cells producing predominantly GM-CSF are expanded in synovial tissues from patients with spondyloarthritis. GM-CSF+CD4+ cells, isolated using a triple cytokine capture approach, have a specific transcriptional signature. Both GM-CSF+ and IL-17A+GM-CSF+ double-producing CD4 T cells express increased levels of GPR65, a proton-sensing receptor associated with spondyloarthritis in genome-wide association studies and pathogenicity in murine inflammatory disease models. Silencing GPR65 in primary CD4 T cells reduces GM-CSF production. GM-CSF and GPR65 may thus serve as targets for therapeutic intervention of spondyloarthritis.
Conflict of interest statement
Paul Bowness has received research grant funding from the Merck Research Laboratories and Celgene. Mohammad Hussein Al-Mossawi has received unrestricted research grant funding from UCB Pharma. The remaining authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

