Integrating proteomics with electrochemistry for identifying kinase biomarkers
- PMID: 29142712
- PMCID: PMC5667508
- DOI: 10.1039/c5sc00560d
Integrating proteomics with electrochemistry for identifying kinase biomarkers
Abstract
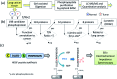
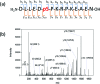
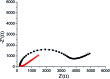
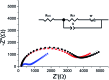
We present an integrated approach for highly sensitive identification and validation of substrate-specific kinases as cancer biomarkers. Our approach combines phosphoproteomics for high throughput cancer-related biomarker discovery from patient tissues and an impedimetric kinase activity biosensor for sensitive validation. Using non-small-cell lung cancer (NSCLC) as a proof-of-concept study, label-free quantitative phosphoproteomic analysis of a pair of cancerous and its adjacent normal tissues revealed 198 phosphoproteins that are over-phosphorylated in NSCLC. Among the differentially regulated phosphorylation sites, the most significant alteration was in residue S165 in the Hepatoma Derived Growth Factor (HDGF) protein. Hence, HDGF was selected as a model system for the electrochemical studies. Further motif-based analysis of this altered phosphorylation site revealed that extracellular-signal-regulated kinase 1/2 (ERK1/2) are most likely to be the corresponding kinases. For validation of the kinase-substrate pair, densely packed peptide monolayers corresponding to the HDGF phosphorylation site were coupled to a gold electrode. Phosphorylation of the monolayer by ERK2 and dephosphorylation by alkaline phosphatase (AP) were detected by electrochemical impedance spectroscopy (EIS) and surface roughness analysis. Compared to other methods for quantification of kinase concentration, this label-free electrochemical assay offers the advantages of ultra-sensitivity as well as higher specificity for the detection of cancer-related kinase-substrate pair. With implementation of multiple kinase-substrate biomarker pairs, we expect this integrated approach to become a high throughput platform for discovery and validation of phosphorylation-mediated biomarkers.
Figures



References
-
- Besant P. G., Tan E., Attwood P. V. Int. J. Biochem. Cell Biol. 2003;35:297–309. - PubMed
-
- Roberts P. J., Der C. J. Oncogene. 2007;26:3291–3310. - PubMed
-
- Ye Y., Zhou X., Li X., Tang Y., Sun Y., Fang J. Tumor Biol. 2014;35:10891–10896. - PubMed
-
- Jerjees D. A., Alabdullah M., Alkaabi M., Abduljabbar R., Muftah A., Nolan C., Green A. R., Ellis I. O., Rakha E. A. Breast Cancer Res. Treat. 2014;147:25–37. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

