Features of Ppd-B1 expression regulation and their impact on the flowering time of wheat near-isogenic lines
- PMID: 29143607
- PMCID: PMC5688470
- DOI: 10.1186/s12870-017-1126-z
Features of Ppd-B1 expression regulation and their impact on the flowering time of wheat near-isogenic lines
Abstract
Background: Photoperiod insensitive Ppd-1a alleles determine early flowering of wheat. Increased expression of homoeologous Ppd-D1a and Ppd-A1a result from deletions in the promoter region, and elevated expression of Ppd-B1a is determined by an increased copy number.
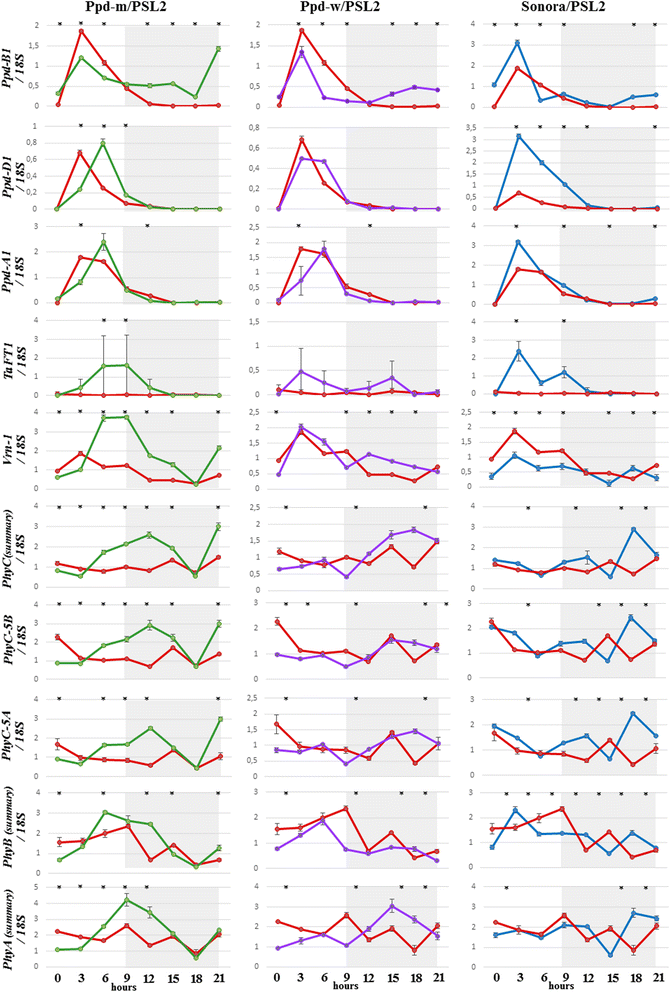
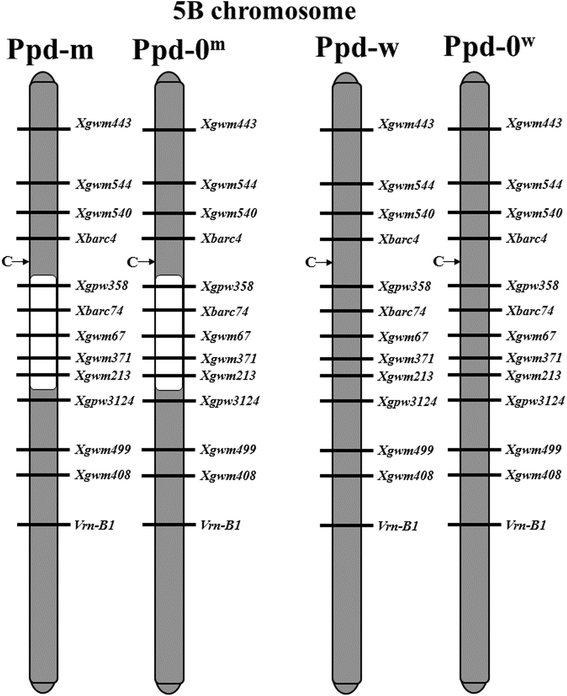
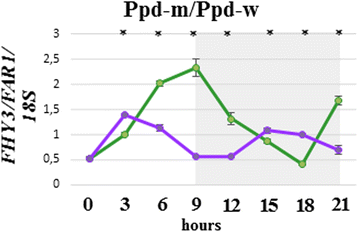
Results: In this study, using bread wheat cultivars Sonora and PSL2, which contrast in flowering time, and near-isogenic lines resulting from their cross, "Ppd-m" and "Ppd-w" with Ppd-B1a introgressed from Sonora, we investigated the putative factors that influence Ppd-B1a expression. By analyzing the Ppd-B1a three distinct copies, we identified an indel and the two SNPs, which distinguished the investigated allele from other alleles with a copy number variation. We studied the expression of the Ppd-A1, Ppd-B1a, and Ppd-D1 genes along with genes that are involved in light perception (PhyA, PhyB, PhyC) and the flowering initiation (Vrn-1, TaFT1) and discussed their interactions. Expression of Ppd-B1a in the "Ppd-m" line, which flowered four days earlier than "Ppd-w", was significantly higher. We found PhyC to be up-regulated in lines with Ppd-B1a alleles. Expression of PhyC was higher in "Ppd-m". Microsatellite genotyping demonstrated that in the line "Ppd-m", there is an introgression in the pericentromeric region of chromosome 5B from the early flowering parental Sonora, while the "Ppd-w" does not have this introgression. FHY3/FAR1 is known to be located in this region. Expression of the transcription factor FHY3/FAR1 was higher in the "Ppd-m" line than in "Ppd-w", suggesting that FHY3/FAR1 is important for the wheat flowering time and may cause earlier flowering of "Ppd-m" as compared to "Ppd-w".
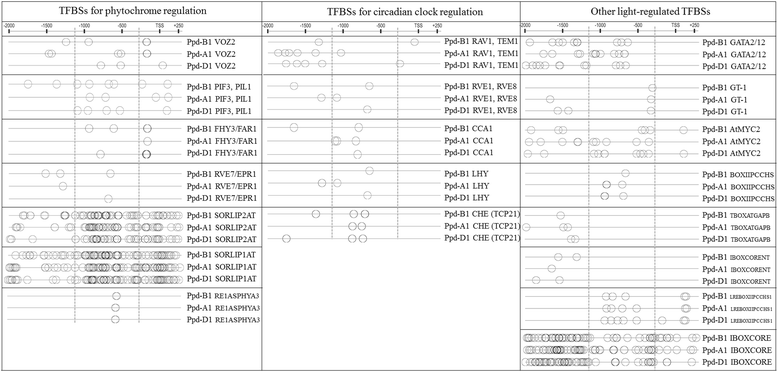
Conclusions: We propose that there is a positive bidirectional regulation of Ppd-B1a and PhyC with an FHY3/FAR1 contribution. The bidirectional regulation can be proposed for Ppd-A1a and Ppd-D1a. Using in silico analysis, we demonstrated that the specificity of the Ppd-B1 regulation compared to that of homoeologous genes involves not only a copy number variation but also distinct regulatory elements.
Keywords: Common wheat; Flowering time; Photoperiod sensitivity; Phytochrome; Ppd-B1.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

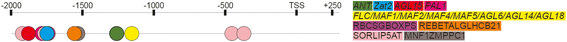
Similar articles
-
Genetics of flowering time in bread wheat Triticum aestivum: complementary interaction between vernalization-insensitive and photoperiod-insensitive mutations imparts very early flowering habit to spring wheat.J Genet. 2012;91(1):33-47. doi: 10.1007/s12041-012-0149-3. J Genet. 2012. PMID: 22546824
-
Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China.BMC Plant Biol. 2013 Dec 5;13:199. doi: 10.1186/1471-2229-13-199. BMC Plant Biol. 2013. PMID: 24314021 Free PMC article.
-
Interactive effects of multiple vernalization (Vrn-1)- and photoperiod (Ppd-1)-related genes on the growth habit of bread wheat and their association with heading and flowering time.BMC Plant Biol. 2018 Dec 27;18(1):374. doi: 10.1186/s12870-018-1587-8. BMC Plant Biol. 2018. PMID: 30587132 Free PMC article.
-
Genome-wide association study of heading and flowering dates and construction of its prediction equation in Chinese common wheat.Theor Appl Genet. 2018 Nov;131(11):2271-2285. doi: 10.1007/s00122-018-3181-8. Epub 2018 Sep 14. Theor Appl Genet. 2018. PMID: 30218294 Review.
-
Major flowering time genes of barley: allelic diversity, effects, and comparison with wheat.Theor Appl Genet. 2021 Jul;134(7):1867-1897. doi: 10.1007/s00122-021-03824-z. Epub 2021 May 9. Theor Appl Genet. 2021. PMID: 33969431 Free PMC article. Review.
Cited by
-
Effects of Rht17 in combination with Vrn-B1 and Ppd-D1 alleles on agronomic traits in wheat in black earth and non-black earth regions.BMC Plant Biol. 2020 Oct 14;20(Suppl 1):304. doi: 10.1186/s12870-020-02514-0. BMC Plant Biol. 2020. PMID: 33050878 Free PMC article.
-
Novel PHOTOPERIOD-1 gene variants associate with yield-related and root-angle traits in European bread wheat.Theor Appl Genet. 2024 May 10;137(6):125. doi: 10.1007/s00122-024-04634-9. Theor Appl Genet. 2024. PMID: 38727862 Free PMC article.
-
Genome-Wide Identification and Functional Characterization of FAR1-RELATED SEQUENCE (FRS) Family Members in Potato (Solanum tuberosum).Plants (Basel). 2023 Jul 7;12(13):2575. doi: 10.3390/plants12132575. Plants (Basel). 2023. PMID: 37447143 Free PMC article.
-
FAR1/FHY3 Transcription Factors Positively Regulate the Salt and Temperature Stress Responses in Eucalyptus grandis.Front Plant Sci. 2022 May 4;13:883654. doi: 10.3389/fpls.2022.883654. eCollection 2022. Front Plant Sci. 2022. PMID: 35599891 Free PMC article.
-
Development of breeder chip for gene detection and molecular-assisted selection by target sequencing in wheat.Mol Breed. 2023 Feb 9;43(2):13. doi: 10.1007/s11032-023-01359-3. eCollection 2023 Feb. Mol Breed. 2023. PMID: 37313130 Free PMC article.
References
-
- Law CN, Sutka J, Worland AJA. Genetic study of day-length response in wheat. Heredity (Edinb). 1978;41:185–191. doi: 10.1038/hdy.1978.87. - DOI
-
- Scarth R, Law CN. The location of the photoperiod gene, Ppd2 and an additional genetic factor for ear-emergence time on chromosome 2B of wheat. Heredity (Edinb) 1983;51:607–619. doi: 10.1038/hdy.1983.73. - DOI
-
- Worland T, Snape JW. Genetic basis of worldwide wheat varietal improvement. In: William A, Alain B, Maarten G, editors. World wheat book, a history of wheat breeding. Paris: Lavoisier Publishing; 2001. pp. 61–67.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

