The Non-Homologous End Joining Protein PAXX Acts to Restrict HSV-1 Infection
- PMID: 29144403
- PMCID: PMC5707549
- DOI: 10.3390/v9110342
The Non-Homologous End Joining Protein PAXX Acts to Restrict HSV-1 Infection
Abstract
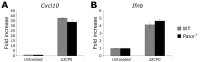
Herpes simplex virus 1 (HSV-1) has extensive interactions with the host DNA damage response (DDR) machinery that can be either detrimental or beneficial to the virus. Proteins in the homologous recombination pathway are known to be required for efficient replication of the viral genome, while different members of the classical non-homologous end-joining (c-NHEJ) pathway have opposing effects on HSV-1 infection. Here, we have investigated the role of the recently-discovered c-NHEJ component, PAXX (Paralogue of XRCC4 and XLF), which we found to be excluded from the nucleus during HSV-1 infection. We have established that cells lacking PAXX have an intact innate immune response to HSV-1 but show a defect in viral genome replication efficiency. Counterintuitively, PAXX-/- cells were able to produce greater numbers of infectious virions, indicating that PAXX acts to restrict HSV-1 infection in a manner that is different from other c-NHEJ factors.
Keywords: DDR; DNA damage response; HSV-1; PAXX; c-NHEJ; classical non-homologous end joining; herpes simplex virus 1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Mimori T., Hardin J.A. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986;261:10375–10379. - PubMed
-
- Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M.C., Sanal O., Plebani A., Stéphan J.L., Hufnagel M., le Deist F., et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

