A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair
- PMID: 29144457
- PMCID: PMC6458054
- DOI: 10.1038/nature24484
A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair
Abstract
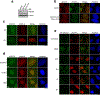
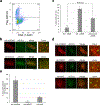
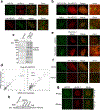
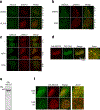
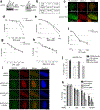
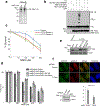
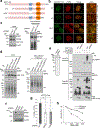
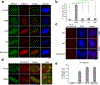
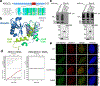
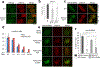
DNA repair is essential to prevent the cytotoxic or mutagenic effects of various types of DNA lesions, which are sensed by distinct pathways to recruit repair factors specific to the damage type. Although biochemical mechanisms for repairing several forms of genomic insults are well understood, the upstream signalling pathways that trigger repair are established for only certain types of damage, such as double-stranded breaks and interstrand crosslinks. Understanding the upstream signalling events that mediate recognition and repair of DNA alkylation damage is particularly important, since alkylation chemotherapy is one of the most widely used systemic modalities for cancer treatment and because environmental chemicals may trigger DNA alkylation. Here we demonstrate that human cells have a previously unrecognized signalling mechanism for sensing damage induced by alkylation. We find that the alkylation repair complex ASCC (activating signal cointegrator complex) relocalizes to distinct nuclear foci specifically upon exposure of cells to alkylating agents. These foci associate with alkylated nucleotides, and coincide spatially with elongating RNA polymerase II and splicing components. Proper recruitment of the repair complex requires recognition of K63-linked polyubiquitin by the CUE (coupling of ubiquitin conjugation to ER degradation) domain of the subunit ASCC2. Loss of this subunit impedes alkylation adduct repair kinetics and increases sensitivity to alkylating agents, but not other forms of DNA damage. We identify RING finger protein 113A (RNF113A) as the E3 ligase responsible for upstream ubiquitin signalling in the ASCC pathway. Cells from patients with X-linked trichothiodystrophy, which harbour a mutation in RNF113A, are defective in ASCC foci formation and are hypersensitive to alkylating agents. Together, our work reveals a previously unrecognized ubiquitin-dependent pathway induced specifically to repair alkylation damage, shedding light on the molecular mechanism of X-linked trichothiodystrophy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

