Antigenicity of the 2015-2016 seasonal H1N1 human influenza virus HA and NA proteins
- PMID: 29145498
- PMCID: PMC5690631
- DOI: 10.1371/journal.pone.0188267
Antigenicity of the 2015-2016 seasonal H1N1 human influenza virus HA and NA proteins
Abstract
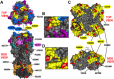
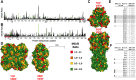
Antigenic drift of the hemagglutinin (HA) and neuraminidase (NA) influenza virus proteins contributes to reduced vaccine efficacy. To analyze antigenic drift in human seasonal H1N1 viruses derived from the 2009 pandemic H1N1 virus (pH1N1-like viruses) accounts for the limited effectiveness (around 40%) of vaccination against pH1N1-like viruses during the 2015-2016 season, nasal washes/swabs collected from adult subjects in the Rochester, NY area, were used to sequence and isolate the circulating viruses. The HA and NA proteins from viruses circulating during the 2015-2016 season encoded eighteen and fourteen amino acid differences, respectively, when compared to A/California/04/2009, a strain circulating at the origin of the 2009 pandemic. The circulating strains belonged to subclade 6B.1, defined by HA amino acid substitutions S101N, S179N, and I233T. Hemagglutination-inhibiting (HAI) and HA-specific neutralizing serum antibody (Ab) titers from around 50% of pH1N1-like virus-infected subjects and immune ferrets were 2-4 fold lower for the 2015-2016 circulating strains compared to the vaccine strain. In addition, using a luminex-based mPlex HA assay, the binding of human sera from subjects infected with pH1N1-like viruses to the HA proteins from circulating and vaccine strains was not identical, strongly suggesting antigenic differences in the HA protein. Additionally, NA inhibition (NAI) Ab titers in human sera from pH1N1-like virus-infected subjects increased after the infection and there were measurable antigenic differences between the NA protein of circulating strains and the vaccine strain using both ferret and human antisera. Despite having been vaccinated, infected subjects exhibited low HAI Ab titers against the vaccine and circulating strains. This suggests that poor responses to the H1N1 component of the vaccine as well as antigenic differences in the HA and NA proteins of currently circulating pH1N1-like viruses could be contributing to risk of infection even after vaccination.
Conflict of interest statement
Figures




Similar articles
-
Antigenic Drift of the Influenza A(H1N1)pdm09 Virus Neuraminidase Results in Reduced Effectiveness of A/California/7/2009 (H1N1pdm09)-Specific Antibodies.mBio. 2019 Apr 9;10(2):e00307-19. doi: 10.1128/mBio.00307-19. mBio. 2019. PMID: 30967460 Free PMC article.
-
Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States.Elife. 2024 Sep 25;13:RP91849. doi: 10.7554/eLife.91849. Elife. 2024. PMID: 39319780 Free PMC article.
-
Antigenic drift of hemagglutinin and neuraminidase in seasonal H1N1 influenza viruses from Saudi Arabia in 2014 to 2015.J Med Virol. 2020 Dec;92(12):3016-3027. doi: 10.1002/jmv.25759. Epub 2020 Apr 1. J Med Virol. 2020. PMID: 32159230 Free PMC article.
-
H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation.Hum Vaccin Immunother. 2018;14(8):1840-1847. doi: 10.1080/21645515.2018.1462639. Epub 2018 May 14. Hum Vaccin Immunother. 2018. PMID: 29641358 Free PMC article. Review.
-
In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen.Viruses. 2014 Jun 23;6(6):2465-94. doi: 10.3390/v6062465. Viruses. 2014. PMID: 24960271 Free PMC article. Review.
Cited by
-
Serologically-Based Evaluation of Cross-Protection Antibody Responses among Different A(H1N1) Influenza Strains.Vaccines (Basel). 2020 Nov 5;8(4):656. doi: 10.3390/vaccines8040656. Vaccines (Basel). 2020. PMID: 33167390 Free PMC article.
-
Immunity to Influenza Infection in Humans.Cold Spring Harb Perspect Med. 2021 Mar 1;11(3):a038729. doi: 10.1101/cshperspect.a038729. Cold Spring Harb Perspect Med. 2021. PMID: 31871226 Free PMC article. Review.
-
Antivirals Targeting the Neuraminidase.Cold Spring Harb Perspect Med. 2022 Jan 4;12(1):a038455. doi: 10.1101/cshperspect.a038455. Cold Spring Harb Perspect Med. 2022. PMID: 32152244 Free PMC article. Review.
-
Repeat vaccination reduces antibody affinity maturation across different influenza vaccine platforms in humans.Nat Commun. 2019 Jul 26;10(1):3338. doi: 10.1038/s41467-019-11296-5. Nat Commun. 2019. PMID: 31350391 Free PMC article. Clinical Trial.
-
A Novel Fluorescent and Bioluminescent Bireporter Influenza A Virus To Evaluate Viral Infections.J Virol. 2019 May 1;93(10):e00032-19. doi: 10.1128/JVI.00032-19. Print 2019 May 15. J Virol. 2019. PMID: 30867298 Free PMC article.
References
-
- Webster RG, Bean WJ Jr. (1978) Genetics of influenza virus. Annu Rev Genet 12: 415–431. doi: 10.1146/annurev.ge.12.120178.002215 - DOI - PubMed
-
- Klenk HD, Garten W (1994) Host cell proteases controlling virus pathogenicity. Trends Microbiol 2: 39–43. - PubMed
-
- Wilson IA, Skehel JJ, Wiley DC (1981) Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289: 366–373. - PubMed
-
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W (1982) The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31: 417–427. - PubMed
-
- Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC (1988) Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333: 426–431. doi: 10.1038/333426a0 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

