The Reactome Pathway Knowledgebase
- PMID: 29145629
- PMCID: PMC5753187
- DOI: 10.1093/nar/gkx1132
The Reactome Pathway Knowledgebase
Abstract
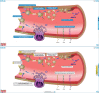
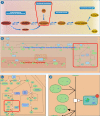
The Reactome Knowledgebase (https://reactome.org) provides molecular details of signal transduction, transport, DNA replication, metabolism, and other cellular processes as an ordered network of molecular transformations-an extended version of a classic metabolic map, in a single consistent data model. Reactome functions both as an archive of biological processes and as a tool for discovering unexpected functional relationships in data such as gene expression profiles or somatic mutation catalogues from tumor cells. To support the continued brisk growth in the size and complexity of Reactome, we have implemented a graph database, improved performance of data analysis tools, and designed new data structures and strategies to boost diagram viewer performance. To make our website more accessible to human users, we have improved pathway display and navigation by implementing interactive Enhanced High Level Diagrams (EHLDs) with an associated icon library, and subpathway highlighting and zooming, in a simplified and reorganized web site with adaptive design. To encourage re-use of our content, we have enabled export of pathway diagrams as 'PowerPoint' files.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A. et al. . The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44:D471–D480. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

