ANAT 2.0: reconstructing functional protein subnetworks
- PMID: 29145805
- PMCID: PMC5689176
- DOI: 10.1186/s12859-017-1932-1
ANAT 2.0: reconstructing functional protein subnetworks
Abstract
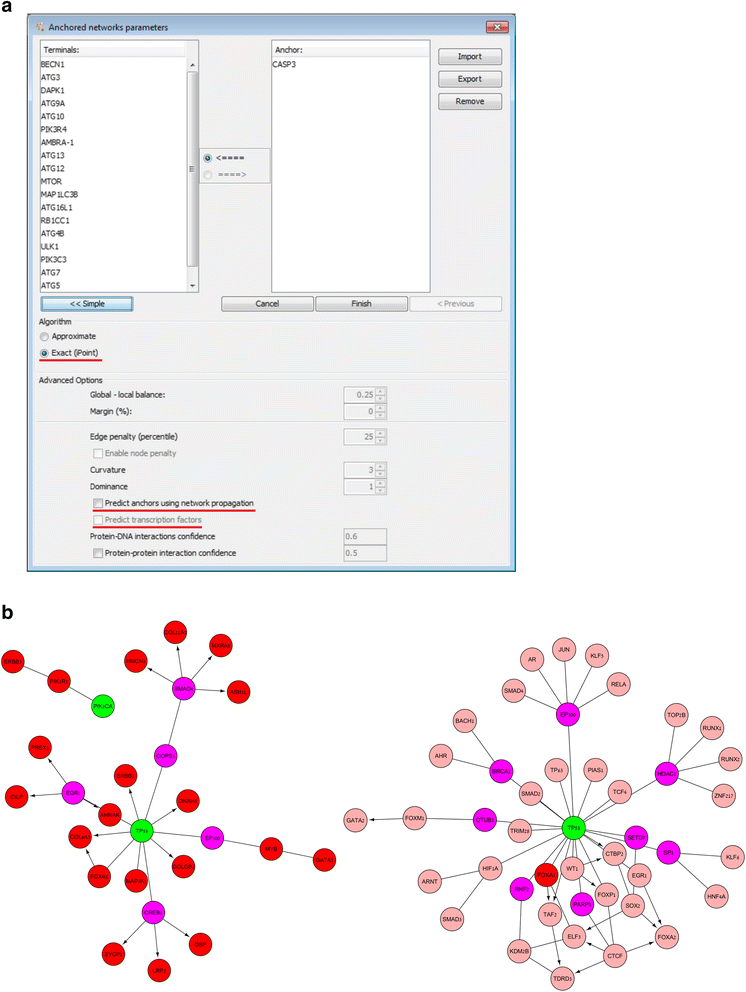
Background: ANAT is a graphical, Cytoscape-based tool for the inference of protein networks that underlie a process of interest. The ANAT tool allows the user to perform network reconstruction under several scenarios in a number of organisms including yeast and human.
Results: Here we report on a new version of the tool, ANAT 2.0, which introduces substantial code and database updates as well as several new network reconstruction algorithms that greatly extend the applicability of the tool to biological data sets.
Conclusions: ANAT 2.0 is an up-to-date network reconstruction tool that addresses several reconstruction challenges across multiple species.
Keywords: Cytoscape plugin; Network inference; Protein-protein interaction network; Subnetwork reconstruction.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

