The Inuit gut microbiome is dynamic over time and shaped by traditional foods
- PMID: 29145891
- PMCID: PMC5689144
- DOI: 10.1186/s40168-017-0370-7
The Inuit gut microbiome is dynamic over time and shaped by traditional foods
Abstract
Background: The human gut microbiome represents a diverse microbial community that varies across individuals and populations, and is influenced by factors such as host genetics and lifestyle. Diet is a major force shaping the gut microbiome, and the effects of dietary choices on microbiome composition are well documented. However, it remains poorly known how natural temporal variation in diet can affect the microbiome. The traditional Inuit diet is primarily based on animal products, which are thought to vary seasonally according to prey availability. We previously investigated the Inuit gut microbiome sampled at a single time point, and found no detectable differences in overall microbiome community composition attributable to the traditional Inuit diet.
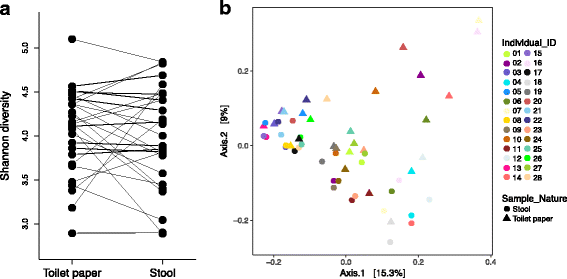
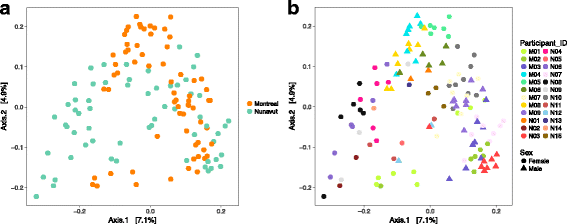
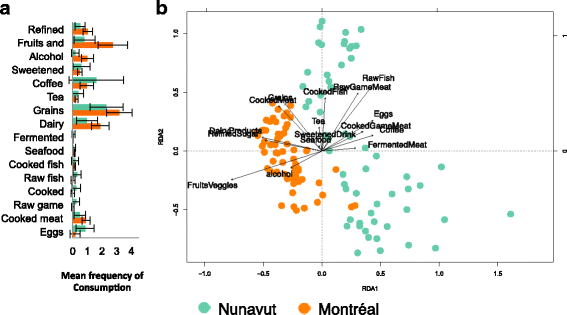
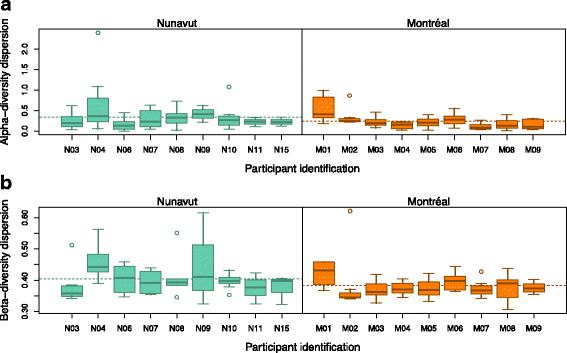
Results: To determine whether seasonal changes in the Inuit diet might induce more pronounced changes in the microbiome, we collected stool and toilet paper samples, and dietary information from Inuit volunteers living in Resolute Bay (Nunavut, Canada), and compared them to individuals of European descent living in Montréal (Québec, Canada) consuming a typical Western diet. We sequenced the V4 region of the 16S rRNA gene to characterize microbiome diversity and composition, and compared samples collected with toilet paper or from stool. Our results show that these sampling methods provide similar, but non-identical portraits of the microbiome. Based on toilet paper samples, we found that much of the variation in microbiome community composition could be explained by individual identity (45-61% of variation explained, depending on the beta diversity metric used), with small but significant variation (3-5%) explained by sex or geography (Nunavut or Montréal). In contrast with our previous study at one time point, sampling over the course of a year revealed that diet explains 11% of variation in community composition across all participants, and 17% of variation specifically among Nunavut participants. However, we observed no clear seasonal shifts in the microbiomes of participants from either Nunavut or Montréal. Within-individual microbial diversity fluctuated more over time in Nunavut than in Montréal, consistent with a more variable and highly individualized diet in Nunavut.
Conclusions: Together, these results shows that the traditional Inuit diet and lifestyle has an impact on the composition, diversity and stability of the Inuit gut microbiome, even if the seasonality of the diet is less pronounced than expected, perhaps due to an increasingly westernized diet.
Keywords: 16S rRNA gene; Dietary transition; Gut microbiome; Inuit traditional diet; Temporal variation; Western diet.
Conflict of interest statement
Ethics approval and consent to participate
Prior to the sample collection, recruitment and sampling protocols were approved by the Université de Montréal Faculty of Arts and Sciences ethical review board (CERFAS, certificate # 2015–16-039-D). Permission for this work was granted by the Nunavut Research Institute (licenses # 02037 15 N–M and # 02040 16 R-M), as well as the Hunters & Trappers Association and the Hamlet of Resolute Bay. Before participating in the study, the objectives and potential outcomes of the research project were explained, and each participant provided written consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

