Division Plane Orientation Defects Revealed by a Synthetic Double Mutant Phenotype
- PMID: 29146775
- PMCID: PMC5761783
- DOI: 10.1104/pp.17.01075
Division Plane Orientation Defects Revealed by a Synthetic Double Mutant Phenotype
Abstract
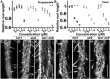
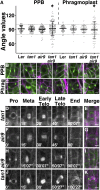
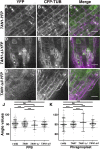
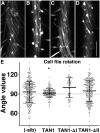
TANGLED1 (TAN1) and AUXIN-INDUCED-IN-ROOTS9 (AIR9) are microtubule-binding proteins that localize to the division site in plants. Their function in Arabidopsis (Arabidopsis thaliana) remained unclear because neither tan1 nor air9 single mutants have a strong phenotype. We show that tan1 air9 double mutants have a synthetic phenotype consisting of short, twisted roots with disordered cortical microtubule arrays that are hypersensitive to a microtubule-depolymerizing drug. The tan1 air9 double mutants have significant defects in division plane orientation due to failures in placing the new cell wall at the correct division site. Full-length TAN1 fused to yellow fluorescent protein, TAN1-YFP, and several deletion constructs were transformed into the double mutant to assess which regions of TAN1 are required for its function in root growth, root twisting, and division plane orientation. TAN1-YFP expressed in tan1 air9 significantly rescued the double mutant phenotype in all three respects. Interestingly, TAN1 missing the first 126 amino acids, TAN1-ΔI-YFP, failed to rescue the double mutant phenotype, while TAN1 missing a conserved middle region, TAN1-ΔII-YFP, significantly rescued the mutant phenotype in terms of root growth and division plane orientation but not root twisting. We use the tan1 air9 double mutant to discover new functions for TAN1 and AIR9 during phragmoplast guidance and root morphogenesis.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Abrash EB, Bergmann DC (2009) Asymmetric cell divisions: a view from plant development. Dev Cell 16: 783–796 - PubMed
-
- Ambrose JC, Cyr R (2008) Mitotic spindle organization by the preprophase band. Mol Plant 1: 950–960 - PubMed
-
- Baskin TI. (2001) On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215: 150–171 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

