Alternative transcription start site selection in Mr-OPY2 controls lifestyle transitions in the fungus Metarhizium robertsii
- PMID: 29146899
- PMCID: PMC5691130
- DOI: 10.1038/s41467-017-01756-1
Alternative transcription start site selection in Mr-OPY2 controls lifestyle transitions in the fungus Metarhizium robertsii
Abstract
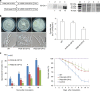
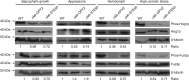
Metarhizium robertsii is a versatile fungus with saprophytic, plant symbiotic and insect pathogenic lifestyle options. Here we show that M. robertsii mediates the saprophyte-to-insect pathogen transition through modulation of the expression of a membrane protein, Mr-OPY2. Abundant Mr-OPY2 protein initiates appressorium formation, a prerequisite for infection, whereas reduced production of Mr-OPY2 elicits saprophytic growth and conidiation. The precise regulation of Mr-OPY2 protein production is achieved via alternative transcription start sites. During saprophytic growth, a single long transcript is produced with small upstream open reading frames in its 5' untranslated region. Increased production of Mr-OPY2 protein on host cuticle is achieved by expression of a transcript variant lacking a small upstream open reading frame that would otherwise inhibit translation of Mr-OPY2. RNA-seq and qRT-PCR analyses show that Mr-OPY2 is a negative regulator of a transcription factor that we demonstrate is necessary for appressorial formation. These findings provide insights into the mechanisms regulating fungal lifestyle transitions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

