LRPPRC-mediated folding of the mitochondrial transcriptome
- PMID: 29146908
- PMCID: PMC5691074
- DOI: 10.1038/s41467-017-01221-z
LRPPRC-mediated folding of the mitochondrial transcriptome
Abstract
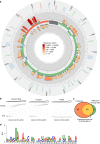
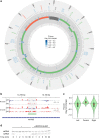
The expression of the compact mammalian mitochondrial genome requires transcription, RNA processing, translation and RNA decay, much like the more complex chromosomal systems, and here we use it as a model system to understand the fundamental aspects of gene expression. Here we combine RNase footprinting with PAR-CLIP at unprecedented depth to reveal the importance of RNA-protein interactions in dictating RNA folding within the mitochondrial transcriptome. We show that LRPPRC, in complex with its protein partner SLIRP, binds throughout the mitochondrial transcriptome, with a preference for mRNAs, and its loss affects the entire secondary structure and stability of the transcriptome. We demonstrate that the LRPPRC-SLIRP complex is a global RNA chaperone that stabilizes RNA structures to expose the required sites for translation, stabilization, and polyadenylation. Our findings reveal a general mechanism where extensive RNA-protein interactions ensure that RNA is accessible for its biological functions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

