The transcription factor Batf3 inhibits the differentiation of regulatory T cells in the periphery
- PMID: 29147008
- PMCID: PMC5704186
- DOI: 10.1038/emm.2017.157
The transcription factor Batf3 inhibits the differentiation of regulatory T cells in the periphery
Abstract
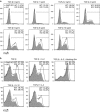
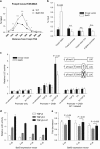
Naive CD4 T cells activated by antigen-presenting cells (APCs) undergo terminal differentiation in the periphery. Multiple mechanisms determine their fates, that is, whether they differentiate into conventional T (Tconv) cells or regulatory T (Treg) cells. The key event during Treg generation is expression of the transcription factor Foxp3, which is the lineage-determining regulator for Treg differentiation and function. Here we show that the transcription factor Batf3 acts as a fate-decision factor with respect to Tconv versus Tregs by restraining Treg differentiation. Batf3 was preferentially expressed in effector CD4 T cells but not in Treg cells, and ectopic expression of Batf3 inhibited Foxp3 induction. Batf3-deficient CD4 T cells favorably differentiated into Treg cells in vitro and in colonic lamina propria. Batf3 KO mice also showed enhanced Treg function in gut-associated immune disease models (for example, ovalbumin tolerance and inflammatory bowel disease models). Batf3 bound to the CNS1 region of the Foxp3 locus and reduced expression of the gene. Thus, Batf3 is a transcriptional suppressor of Treg differentiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
OX40 Costimulation Inhibits Foxp3 Expression and Treg Induction via BATF3-Dependent and Independent Mechanisms.Cell Rep. 2018 Jul 17;24(3):607-618. doi: 10.1016/j.celrep.2018.06.052. Cell Rep. 2018. PMID: 30021159 Free PMC article.
-
Egress of mature murine regulatory T cells from the thymus requires RelA.J Immunol. 2015 Apr 1;194(7):3020-8. doi: 10.4049/jimmunol.1302756. Epub 2015 Feb 27. J Immunol. 2015. PMID: 25725099
-
Dendritic Cells Coordinate the Development and Homeostasis of Organ-Specific Regulatory T Cells.Immunity. 2016 Apr 19;44(4):847-59. doi: 10.1016/j.immuni.2016.01.025. Epub 2016 Mar 29. Immunity. 2016. PMID: 27037189 Free PMC article.
-
Differentiation and function of Foxp3(+) effector regulatory T cells.Trends Immunol. 2013 Feb;34(2):74-80. doi: 10.1016/j.it.2012.11.002. Epub 2012 Dec 6. Trends Immunol. 2013. PMID: 23219401 Review.
-
Distinct regulatory CD4+T cell subsets; differences between naïve and antigen specific T regulatory cells.Curr Opin Immunol. 2011 Oct;23(5):641-7. doi: 10.1016/j.coi.2011.07.012. Epub 2011 Aug 11. Curr Opin Immunol. 2011. PMID: 21840184 Review.
Cited by
-
Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes.Nat Immunol. 2020 May;21(5):578-587. doi: 10.1038/s41590-020-0633-5. Epub 2020 Mar 30. Nat Immunol. 2020. PMID: 32231298 Free PMC article.
-
The Balance of Th17 versus Treg Cells in Autoimmunity.Int J Mol Sci. 2018 Mar 3;19(3):730. doi: 10.3390/ijms19030730. Int J Mol Sci. 2018. PMID: 29510522 Free PMC article. Review.
-
Targeting the Dendritic Cell-Secreted Immunoregulatory Cytokine CCL22 Alleviates Radioresistance.Clin Cancer Res. 2024 Oct 1;30(19):4450-4463. doi: 10.1158/1078-0432.CCR-23-3616. Clin Cancer Res. 2024. PMID: 38691100 Free PMC article.
-
Pathway analysis of smoking-induced changes in buccal mucosal gene expression.Egypt J Med Hum Genet. 2022;23(1):69. doi: 10.1186/s43042-022-00268-y. Epub 2022 Mar 17. Egypt J Med Hum Genet. 2022. PMID: 37521848 Free PMC article.
-
Single cell sequencing reveals that CD39 inhibition mediates changes to the tumor microenvironment.Nat Commun. 2022 Nov 8;13(1):6740. doi: 10.1038/s41467-022-34495-z. Nat Commun. 2022. PMID: 36347860 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

