A G3P[13] porcine group A rotavirus emerging in China is a reassortant and a natural recombinant in the VP4 gene
- PMID: 29148270
- PMCID: PMC7169750
- DOI: 10.1111/tbed.12756
A G3P[13] porcine group A rotavirus emerging in China is a reassortant and a natural recombinant in the VP4 gene
Abstract
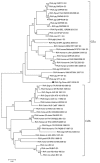
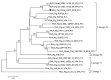
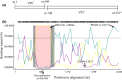
Group A rotaviruses (RVAs) are a major cause of serious intestinal disease in piglets. In this study, a novel pig strain was identified in a stool sample from China. The strain was designated RVA/Pig/China/LNCY/2016/G3P[13] and had a G3-P[13]-I5-R1-C1-M1-A8-N1-T1-E1-H1 genome. The viral protein 7 (VP7) and non-structural protein 4 (NSP4) genes of RVA/Pig/China/LNCY/2016/G3P[13] were closely related to cogent genes of human RVAs, suggesting that a reassortment between pig and human strains had occurred. Recombination analysis showed that RVA/Pig/China/LNCY/2016/G3P[13] is a natural recombinant strain between the P[23] and P[7] RVA strains, and crossover points for recombination were found at nucleotides (nt) 456 and 804 of the VP4 gene. Elucidating the biological characteristics of porcine rotavirus (PoRV) will be helpful for further analyses of the epidemic characteristics of this virus. The results of this study provide valuable information for RVA recombination and evolution and will facilitate future investigations into the molecular pathogenesis of RVAs.
Keywords: VP4; porcine group A rotavirus; reassortant; recombinant.
© 2017 Blackwell Verlag GmbH.
Figures


References
-
- Amimo, J. O. , Vlasova, A. N. , & Saif, L. J. (2013). Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011 and 2012) swine fecal samples in Ohio: Predominance of the G9P[13] genotype in nursing piglets. Journal of Clinical Microbiology, 51, 1142–1151. 10.1128/JCM.03193-12 - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

