Molecular Alliance of Lymantria dispar Multiple Nucleopolyhedrovirus and a Short Unmodified Antisense Oligonucleotide of Its Anti-Apoptotic IAP-3 Gene: A Novel Approach for Gypsy Moth Control
- PMID: 29149051
- PMCID: PMC5713413
- DOI: 10.3390/ijms18112446
Molecular Alliance of Lymantria dispar Multiple Nucleopolyhedrovirus and a Short Unmodified Antisense Oligonucleotide of Its Anti-Apoptotic IAP-3 Gene: A Novel Approach for Gypsy Moth Control
Abstract
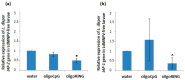
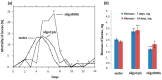
Baculovirus IAP (inhibitor-of-apoptosis) genes originated by capture of host genes. Unmodified short antisense DNA oligonucleotides (oligoDNAs) from baculovirus IAP genes can down-regulate specific gene expression profiles in both baculovirus-free and baculovirus-infected insects. In this study, gypsy moth (Lymantria dispar) larvae infected with multiple nucleopolyhedrovirus (LdMNPV), and LdMNPV-free larvae, were treated with oligoDNA antisense to the RING (really interesting new gene) domain of the LdMNPV IAP-3 gene. The results with respect to insect mortality, biomass accumulation, histological studies, RT-PCR, and analysis of DNA apoptotic fragmentation suggest that oligoRING induced increased apoptotic processes in both LdMNPV-free and LdMNPV-infected insect cells, but were more pronounced in the latter. These data open up possibilities for promising new routes of insect pest control using antisense phosphodiester DNA oligonucleotides.
Keywords: DNA insecticides; RING (really interesting new gene); baculovirus infection; gypsy moth Lymantria dispar; insect pest management; multiple nucleopolyhedrovirus IAP (inhibitor-of-apoptosis) genes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Gu L., Knipple D.C. Recent advances in RNA interference research in insects: Implications for future insect pest management strategies. Crop. Protect. 2013;45:36–40. doi: 10.1016/j.cropro.2012.10.004. - DOI
-
- Terenius O., Papanicolaou A., Garbutt J.S., Eleftherianos I., Huvenne H., Kanginakudru S., Albrechtsen M., An C., Aymeric J.L., Barthel A., et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. - DOI - PubMed
-
- Lundgren J.G., Duan J.J. RNAi-based insecticidal crops: Potential effects on nontargetspecies. Bioscience. 2013;63:657–665. doi: 10.1525/bio.2013.63.8.8. - DOI
-
- Oberemok V.V. Method of elimination of phyllophagousinsects from order Lepidoptera. 36445. UA Patent. 2008 May 19;
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

