Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species
- PMID: 29149178
- PMCID: PMC5711037
- DOI: 10.1371/journal.pbio.2003583
Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species
Abstract
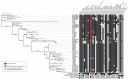
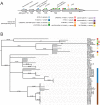
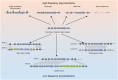
Filamentous fungi produce a diverse array of secondary metabolites (SMs) critical for defense, virulence, and communication. The metabolic pathways that produce SMs are found in contiguous gene clusters in fungal genomes, an atypical arrangement for metabolic pathways in other eukaryotes. Comparative studies of filamentous fungal species have shown that SM gene clusters are often either highly divergent or uniquely present in one or a handful of species, hampering efforts to determine the genetic basis and evolutionary drivers of SM gene cluster divergence. Here, we examined SM variation in 66 cosmopolitan strains of a single species, the opportunistic human pathogen Aspergillus fumigatus. Investigation of genome-wide within-species variation revealed 5 general types of variation in SM gene clusters: nonfunctional gene polymorphisms; gene gain and loss polymorphisms; whole cluster gain and loss polymorphisms; allelic polymorphisms, in which different alleles corresponded to distinct, nonhomologous clusters; and location polymorphisms, in which a cluster was found to differ in its genomic location across strains. These polymorphisms affect the function of representative A. fumigatus SM gene clusters, such as those involved in the production of gliotoxin, fumigaclavine, and helvolic acid as well as the function of clusters with undefined products. In addition to enabling the identification of polymorphisms, the detection of which requires extensive genome-wide synteny conservation (e.g., mobile gene clusters and nonhomologous cluster alleles), our approach also implicated multiple underlying genetic drivers, including point mutations, recombination, and genomic deletion and insertion events as well as horizontal gene transfer from distant fungi. Finally, most of the variants that we uncover within A. fumigatus have been previously hypothesized to contribute to SM gene cluster diversity across entire fungal classes and phyla. We suggest that the drivers of genetic diversity operating within a fungal species shown here are sufficient to explain SM cluster macroevolutionary patterns.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Vining LC. Functions of secondary metabolites. Annu Rev Microbiol. Annual Reviews 4139 El Camino Way, P.O. Box 10139, Palo Alto, CA 94303–0139, USA; 1990;44: 395–427. doi: 10.1146/annurev.mi.44.100190.002143 - DOI - PubMed
-
- Schimek C. Evolution of Special Metabolism in Fungi: Concepts, Mechanisms, and Pathways In: Pöggler S, Wöstmeyer J, editors. Evolution of Fungi and Fungal-Like Organisms, The Mycota. XIV Berlin, Heidelberg: Springer-Verlag; 2011. pp. 293–328.
-
- Fox EM, Howlett BJ. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 2008;11: 481–7. doi: 10.1016/j.mib.2008.10.007 - DOI - PubMed
-
- Scharf DH, Heinekamp T, Brakhage AA. Human and Plant Fungal Pathogens: The Role of Secondary Metabolites. PLoS Pathog. 2014;10(1): e1003859 doi: 10.1371/journal.ppat.1003859 - DOI - PMC - PubMed
-
- Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci. 2007;362: 1195–200. doi: 10.1098/rstb.2007.2044 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

