The Effect of Nonreversibility on Inferring Rooted Phylogenies
- PMID: 29149300
- PMCID: PMC5889004
- DOI: 10.1093/molbev/msx294
The Effect of Nonreversibility on Inferring Rooted Phylogenies
Erratum in
-
Molecular Biology and Evolution, Volume 35, Issue 4.Mol Biol Evol. 2018 Jul 1;35(7):1821. doi: 10.1093/molbev/msy057. Mol Biol Evol. 2018. PMID: 29659984 Free PMC article. No abstract available.
Abstract
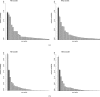
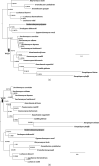
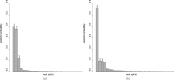
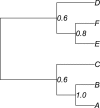
Most phylogenetic models assume that the evolutionary process is stationary and reversible. In addition to being biologically improbable, these assumptions also impair inference by generating models under which the likelihood does not depend on the position of the root. Consequently, the root of the tree cannot be inferred as part of the analysis. Yet identifying the root position is a key component of phylogenetic inference because it provides a point of reference for polarizing ancestor-descendant relationships and therefore interpreting the tree. In this paper, we investigate the effect of relaxing the unrealistic reversibility assumption and allowing the position of the root to be another unknown. We propose two hierarchical models that are centered on a reversible model but perturbed to allow nonreversibility. The models differ in the degree of structure imposed on the perturbations. The analysis is performed in the Bayesian framework using Markov chain Monte Carlo methods for which software is provided. We illustrate the performance of the two nonreversible models in analyses of simulated data using two types of topological priors. We then apply the models to a real biological data set, the radiation of polyploid yeasts, for which there is robust biological opinion about the root position. Finally, we apply the models to a second biological alignment for which the rooted tree is controversial: the ribosomal tree of life. We compare the two nonreversible models and conclude that both are useful in inferring the position of the root from real biological data.
Figures




References
-
- Alfaro ME, Holder MT.. 2006. The posterior and the prior in Bayesian phylogenetics. Annu Rev Ecol Evol Syst. 371:19–42.10.1146/annurev.ecolsys.37.091305.110021 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

