Binding Thermodynamics and Kinetics Calculations Using Chemical Host and Guest: A Comprehensive Picture of Molecular Recognition
- PMID: 29149564
- PMCID: PMC5920803
- DOI: 10.1021/acs.jctc.7b00899
Binding Thermodynamics and Kinetics Calculations Using Chemical Host and Guest: A Comprehensive Picture of Molecular Recognition
Abstract
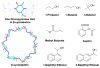
Understanding the fine balance between changes of entropy and enthalpy and the competition between a guest and water molecules in molecular binding is crucial in fundamental studies and practical applications. Experiments provide measurements. However, illustrating the binding/unbinding processes gives a complete picture of molecular recognition not directly available from experiments, and computational methods bridge the gaps. Here, we investigated guest association/dissociation with β-cyclodextrin (β-CD) by using microsecond-time-scale molecular dynamics (MD) simulations, postanalysis and numerical calculations. We computed association and dissociation rate constants, enthalpy, and solvent and solute entropy of binding. All the computed values of kon, koff, ΔH, ΔS, and ΔG using GAFF-CD and q4MD-CD force fields for β-CD could be compared with experimental data directly and agreed reasonably with experiment findings. In addition, our study further interprets experiments. Both force fields resulted in similar computed ΔG from independently computed kinetics rates, ΔG = -RT ln(kon·C0/koff), and thermodynamics properties, ΔG = ΔH - TΔS. The water entropy calculations show that the entropy gain of desolvating water molecules are a major driving force, and both force fields have the same strength of nonpolar attractions between solutes and β-CD as well. Water molecules play a crucial role in guest binding to β-CD. However, collective water/β-CD motions could contribute to different computed kon and ΔH values by different force fields, mainly because the parameters of β-CD provide different motions of β-CD, hydrogen-bond networks of water molecules in the cavity of free β-CD, and strength of desolvation penalty. As a result, q4MD-CD suggests that guest binding is mostly driven by enthalpy, while GAFF-CD shows that gaining entropy is the major driving force of binding. The study deepens our understanding of ligand-receptor recognition and suggests strategies for force field parametrization for accurately modeling molecular systems.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
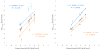
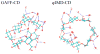


Similar articles
-
Analyses of polycyclic aromatic hydrocarbon (PAH) and chiral-PAH analogues-methyl-β-cyclodextrin guest-host inclusion complexes by fluorescence spectrophotometry and multivariate regression analysis.Spectrochim Acta A Mol Biomol Spectrosc. 2017 Mar 5;174:316-325. doi: 10.1016/j.saa.2016.11.047. Epub 2016 Dec 5. Spectrochim Acta A Mol Biomol Spectrosc. 2017. PMID: 27984752
-
Binding Thermodynamics of Host-Guest Systems with SMIRNOFF99Frosst 1.0.5 from the Open Force Field Initiative.J Chem Theory Comput. 2019 Nov 12;15(11):6225-6242. doi: 10.1021/acs.jctc.9b00748. Epub 2019 Oct 25. J Chem Theory Comput. 2019. PMID: 31603667 Free PMC article.
-
Statistical properties of thermodynamic quantities for cyclodextrin complex formation.J Pharm Sci. 2000 Nov;89(11):1389-94. doi: 10.1002/1520-6017(200011)89:11<1389::aid-jps2>3.0.co;2-m. J Pharm Sci. 2000. PMID: 11015684
-
Can thermodynamic measurements of receptor binding yield information on drug affinity and efficacy?Biochem Pharmacol. 2000 Dec 1;60(11):1549-56. doi: 10.1016/s0006-2952(00)00368-3. Biochem Pharmacol. 2000. PMID: 11077036 Review.
-
Thermodynamics of protein-ligand interactions: history, presence, and future aspects.J Recept Signal Transduct Res. 2004 Feb;24(1-2):1-52. doi: 10.1081/rrs-120037896. J Recept Signal Transduct Res. 2004. PMID: 15344878 Review.
Cited by
-
Ligand Gaussian Accelerated Molecular Dynamics 2 (LiGaMD2): Improved Calculations of Ligand Binding Thermodynamics and Kinetics with Closed Protein Pocket.J Chem Theory Comput. 2023 Feb 14;19(3):733-745. doi: 10.1021/acs.jctc.2c01194. Epub 2023 Jan 27. J Chem Theory Comput. 2023. PMID: 36706316 Free PMC article.
-
Binding Kinetics Toolkit for Analyzing Transient Molecular Conformations and Computing Free Energy Landscapes.J Phys Chem A. 2022 Nov 24;126(46):8761-8770. doi: 10.1021/acs.jpca.2c05499. Epub 2022 Nov 8. J Phys Chem A. 2022. PMID: 36346951 Free PMC article.
-
Enhanced-Sampling Simulations for the Estimation of Ligand Binding Kinetics: Current Status and Perspective.Front Mol Biosci. 2022 Jun 8;9:899805. doi: 10.3389/fmolb.2022.899805. eCollection 2022. Front Mol Biosci. 2022. PMID: 35755817 Free PMC article. Review.
-
SEEKR2: Versatile Multiscale Milestoning Utilizing the OpenMM Molecular Dynamics Engine.J Chem Inf Model. 2022 Jul 11;62(13):3253-3262. doi: 10.1021/acs.jcim.2c00501. Epub 2022 Jun 27. J Chem Inf Model. 2022. PMID: 35759413 Free PMC article.
-
Quantitative Ranking of Ligand Binding Kinetics with a Multiscale Milestoning Simulation Approach.J Phys Chem Lett. 2018 Sep 6;9(17):4941-4948. doi: 10.1021/acs.jpclett.8b02047. Epub 2018 Aug 16. J Phys Chem Lett. 2018. PMID: 30070844 Free PMC article.
References
-
- Leavitt S, Freire E. Direct Measurement of Protein Binding Energetics by Isothermal Titration Calorimetry. Curr Opin Struct Biol. 2001;11(5):560–566. - PubMed
-
- Bouchemal K, Mazzaferro S. How to Conduct and Interpret Itc Experiments Accurately for Cyclodextrin-Guest Interactions. Drug Discovery Today. 2012;17(11–12):623–629. - PubMed
-
- Swinney DC. The Role of Binding Kinetics in Therapeutically Useful Drug Action. Curr Opin Drug Discovery Dev. 2009;12(1):31– 39. - PubMed
-
- Zhang R, Monsma F. The Importance of Drug-Target Residence Time. Curr Opin Drug Discovery Dev. 2009;12(4):488–496. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
