Tissue-Resident Macrophages Are Locally Programmed for Silent Clearance of Apoptotic Cells
- PMID: 29150239
- PMCID: PMC5728676
- DOI: 10.1016/j.immuni.2017.10.006
Tissue-Resident Macrophages Are Locally Programmed for Silent Clearance of Apoptotic Cells
Abstract
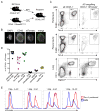
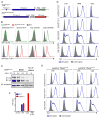
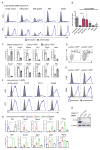
Although apoptotic cells (ACs) contain nucleic acids that can be recognized by Toll-like receptors (TLRs), engulfment of ACs does not initiate inflammation in healthy organisms. Here we identified macrophage populations that continually engulf ACs in distinct tissues and found that these macrophages share characteristics compatible with immunologically silent clearance of ACs; such characteristics include high expression of AC recognition receptors, low expression of TLR9, and reduced TLR responsiveness to nucleic acids. Removal of the macrophages from tissues resulted in loss of many of these characteristics and the ability to generate inflammatory responses to AC-derived nucleic acids, suggesting that cues from the tissue microenvironment program macrophages for silent AC clearance. The transcription factors KLF2 and KLF4 control the expression of many genes within this AC clearance program. The coordinated expression of AC receptors with genes that limit responses to nucleic acids might ensure maintenance of homeostasis and thus represent a central feature of tissue macrophages.
Keywords: KLF2; KLF4; Toll-like receptors; apoptotic cell clearance; autoinflammation; macrophage.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- Baratin M, Simon L, Jorquera A, Ghigo C, Dembele D, Nowak J, Gentek R, Wienert S, Klauschen F, Malissen B, et al. T Cell Zone Resident Macrophages Silently Dispose of Apoptotic Cells in the Lymph Node. Immunity. 2017;47:349–362. e5. - PubMed
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. - PubMed
-
- Barrat FJ, Elkon KB, Fitzgerald KA. Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu Rev Med. 2016;67:323–336. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

