Dysregulation of mRNA translation and energy metabolism in cancer
- PMID: 29150352
- PMCID: PMC5993950
- DOI: 10.1016/j.jbior.2017.11.001
Dysregulation of mRNA translation and energy metabolism in cancer
Abstract
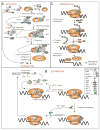
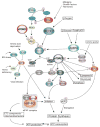
Dysregulated mRNA translation and aberrant energy metabolism are frequent in cancer. Considering that mRNA translation is an energy demanding process, cancer cells must produce sufficient ATP to meet energy demand of hyperactive translational machinery. In recent years, the mammalian/mechanistic target of rapamycin (mTOR) emerged as a central regulatory node which coordinates energy consumption by the translation apparatus and ATP production in mitochondria. Aberrant mTOR signaling underpins the vast majority of cancers whereby increased mTOR activity is thought to be a major determinant of both malignant translatomes and metabolomes. Nonetheless, the role of mTOR and other related signaling nodes (e.g. AMPK) in orchestrating protein synthesis and cancer energetics is only recently being unraveled. In this review, we discuss recent findings that provide insights into the molecular underpinnings of coordination of translational and metabolic programs of cancer cells, and potential strategies to translate these findings into clinical treatments.
Keywords: Cancer; Energy metabolism; mRNA translation; mTOR.
Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Algire C, Amrein L, Bazile M, David S, Zakikhani M, Pollak M. Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene 2010 - PubMed
-
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125(6):1125–1136. - PubMed
-
- Audet-Walsh E, Dufour CR, Yee T, Zouanat FZ, Yan M, Kalloghlian G, Vernier M, Caron M, Bourque G, Scarlata E, Hamel L, Brimo F, Aprikian AG, Lapointe J, Chevalier S, Giguere V. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes & development 2017 - PMC - PubMed
-
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. Architecture of human mTOR complex 1. Science (New York, NY) 2016;351(6268):48–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

