glnA Truncation in Salmonella enterica Results in a Small Colony Variant Phenotype, Attenuated Host Cell Entry, and Reduced Expression of Flagellin and SPI-1-Associated Effector Genes
- PMID: 29150501
- PMCID: PMC5752853
- DOI: 10.1128/AEM.01838-17
glnA Truncation in Salmonella enterica Results in a Small Colony Variant Phenotype, Attenuated Host Cell Entry, and Reduced Expression of Flagellin and SPI-1-Associated Effector Genes
Abstract
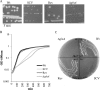
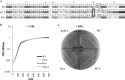
Many pathogenic bacteria use sophisticated survival strategies to overcome harsh environmental conditions. One strategy is the formation of slow-growing subpopulations termed small colony variants (SCVs). Here we characterize an SCV that spontaneously emerged from an axenic Salmonella enterica serovar Typhimurium water culture. We found that the SCV harbored a frameshift mutation in the glutamine synthetase gene glnA, leading to an ∼90% truncation of the corresponding protein. Glutamine synthetase, a central enzyme in nitrogen assimilation, converts glutamate and ammonia to glutamine. Glutamine is an important nitrogen donor that is required for the synthesis of cellular compounds. The internal glutamine pool serves as an indicator of nitrogen availability in Salmonella In our study, the SCV and a constructed glnA knockout mutant showed reduced growth rates, compared to the wild type. Moreover, the SCV and the glnA mutant displayed attenuated entry into host cells and severely reduced levels of exoproteins, including flagellin and several Salmonella pathogenicity island 1 (SPI-1)-dependent secreted virulence factors. We found that these proteins were also depleted in cell lysates, indicating their diminished synthesis. Accordingly, the SCV and the glnA mutant had severely decreased expression of flagellin genes, several SPI-1 effector genes, and a class 2 motility gene (flgB). However, the expression of a class 1 motility gene (flhD) was not affected. Supplementation with glutamine or genetic reversion of the glnA truncation restored growth, cell entry, gene expression, and protein abundance. In summary, our data show that glnA is essential for the growth of S. enterica and controls important motility- and virulence-related traits in response to glutamine availability.IMPORTANCESalmonella enterica serovar Typhimurium is a significant pathogen causing foodborne infections. Here we describe an S Typhimurium small colony variant (SCV) that spontaneously emerged from a long-term starvation experiment in water. It is important to study SCVs because (i) SCVs may arise spontaneously upon exposure to stresses, including environmental and host defense stresses, (ii) SCVs are slow growing and difficult to eradicate, and (iii) only a few descriptions of S. enterica SCVs are available. We clarify the genetic basis of the SCV described here as a frameshift mutation in the glutamine synthetase gene glnA, leading to glutamine auxotrophy. In Salmonella, internal glutamine limitation serves as a sign of external nitrogen deficiency and is thought to regulate cell growth. In addition to exhibiting impaired growth, the SCV showed reduced host cell entry and reduced expression of SPI-1 virulence and flagellin genes.
Keywords: SPI-1; Salmonella enterica; glutamine synthetase; motility; small colony variant.
Copyright © 2018 American Society for Microbiology.
Figures

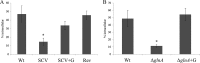


Similar articles
-
Presence of SopE and mode of infection result in increased Salmonella-containing vacuole damage and cytosolic release during host cell infection by Salmonella enterica.Cell Microbiol. 2020 May;22(5):e13155. doi: 10.1111/cmi.13155. Epub 2020 Feb 10. Cell Microbiol. 2020. PMID: 31872936
-
Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions.Curr Biol. 2004 Jun 8;14(11):1018-24. doi: 10.1016/j.cub.2004.05.050. Curr Biol. 2004. PMID: 15182677
-
Salmonella pathogenicity islands in host specificity, host pathogen-interactions and antibiotics resistance of Salmonella enterica.Berl Munch Tierarztl Wochenschr. 2007 Jul-Aug;120(7-8):317-27. Berl Munch Tierarztl Wochenschr. 2007. PMID: 17715824 Review.
-
Salmonella-containing vacuoles display centrifugal movement associated with cell-to-cell transfer in epithelial cells.Infect Immun. 2009 Mar;77(3):996-1007. doi: 10.1128/IAI.01275-08. Epub 2008 Dec 22. Infect Immun. 2009. PMID: 19103768 Free PMC article.
-
Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors.Microbiology (Reading). 2012 May;158(Pt 5):1147-1161. doi: 10.1099/mic.0.058115-0. Epub 2012 Mar 15. Microbiology (Reading). 2012. PMID: 22422755 Review.
Cited by
-
Heliorhodopsin binds and regulates glutamine synthetase activity.PLoS Biol. 2022 Oct 3;20(10):e3001817. doi: 10.1371/journal.pbio.3001817. eCollection 2022 Oct. PLoS Biol. 2022. PMID: 36190943 Free PMC article.
-
Deletion of Glutamine Synthetase Gene Disrupts the Survivability and Infectivity of Leishmania donovani.Front Cell Infect Microbiol. 2021 Feb 26;11:622266. doi: 10.3389/fcimb.2021.622266. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33732662 Free PMC article.
-
Short Chain Fatty Acids Commonly Produced by Gut Microbiota Influence Salmonella enterica Motility, Biofilm Formation, and Gene Expression.Antibiotics (Basel). 2019 Dec 13;8(4):265. doi: 10.3390/antibiotics8040265. Antibiotics (Basel). 2019. PMID: 31847278 Free PMC article.
-
Systematic Evaluation of Whole Genome Sequence-Based Predictions of Salmonella Serotype and Antimicrobial Resistance.Front Microbiol. 2020 Apr 3;11:549. doi: 10.3389/fmicb.2020.00549. eCollection 2020. Front Microbiol. 2020. PMID: 32318038 Free PMC article.
-
Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network.Front Cell Infect Microbiol. 2019 Jul 31;9:270. doi: 10.3389/fcimb.2019.00270. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31428589 Free PMC article. Review.
References
-
- Valdez Y, Ferreira RB, Finlay BB. 2009. Molecular mechanisms of Salmonella virulence and host resistance. Curr Top Microbiol Immunol 337:93–127. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

