Carbon Amendments Alter Microbial Community Structure and Net Mercury Methylation Potential in Sediments
- PMID: 29150503
- PMCID: PMC5772229
- DOI: 10.1128/AEM.01049-17
Carbon Amendments Alter Microbial Community Structure and Net Mercury Methylation Potential in Sediments
Abstract
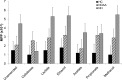
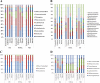
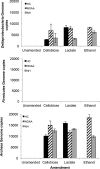
Neurotoxic methylmercury (MeHg) is produced by anaerobic Bacteria and Archaea possessing the genes hgcAB, but it is unknown how organic substrate and electron acceptor availability impacts the distribution and abundance of these organisms. We evaluated the impact of organic substrate amendments on mercury (Hg) methylation rates, microbial community structure, and the distribution of hgcAB+ microbes with sediments. Sediment slurries were amended with short-chain fatty acids, alcohols, or a polysaccharide. Minimal increases in MeHg were observed following lactate, ethanol, and methanol amendments, while a significant decrease (∼70%) was observed with cellobiose incubations. Postincubation, microbial diversity was assessed via 16S rRNA amplicon sequencing. The presence of hgcAB+ organisms was assessed with a broad-range degenerate PCR primer set for both genes, while the presence of microbes in each of the three dominant clades of methylators (Deltaproteobacteria, Firmicutes, and methanogenic Archaea) was measured with clade-specific degenerate hgcA quantitative PCR (qPCR) primer sets. The predominant microorganisms in unamended sediments consisted of Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria Clade-specific qPCR identified hgcA+Deltaproteobacteria and Archaea in all sites but failed to detect hgcA+Firmicutes Cellobiose shifted the communities in all samples to ∼90% non-hgcAB-containing Firmicutes (mainly Bacillus spp. and Clostridium spp.). These results suggest that either expression of hgcAB is downregulated or, more likely given the lack of 16S rRNA gene presence after cellobiose incubation, Hg-methylating organisms are largely outcompeted by cellobiose degraders or degradation products of cellobiose. These results represent a step toward understanding and exploring simple methodologies for controlling MeHg production in the environment.IMPORTANCE Methylmercury (MeHg) is a neurotoxin produced by microorganisms that bioacummulates in the food web and poses a serious health risk to humans. Currently, the impact that organic substrate or electron acceptor availability has on the mercury (Hg)-methylating microorganisms is unclear. To study this, we set up microcosm experiments exposed to different organic substrates and electron acceptors and assayed for Hg methylation rates, for microbial community structure, and for distribution of Hg methylators. The sediment and groundwater was collected from East Fork Poplar Creek in Oak Ridge, TN. Amendment with cellobiose (a lignocellulosic degradation by-product) led to a drastic decrease in the Hg methylation rate compared to that in an unamended control, with an associated shift in the microbial community to mostly nonmethylating Firmicutes This, along with previous Hg-methylating microorganism identification methods, will be important for identifying strategies to control MeHg production and inform future remediation strategies.
Keywords: 16S; hgcA; hgcAB; mercury; methylmercury; qPCR.
Copyright © 2018 Christensen et al.
Figures

Similar articles
-
Development and Validation of Broad-Range Qualitative and Clade-Specific Quantitative Molecular Probes for Assessing Mercury Methylation in the Environment.Appl Environ Microbiol. 2016 Sep 16;82(19):6068-78. doi: 10.1128/AEM.01271-16. Print 2016 Oct 1. Appl Environ Microbiol. 2016. PMID: 27422835 Free PMC article.
-
Periphyton and Flocculent Materials Are Important Ecological Compartments Supporting Abundant and Diverse Mercury Methylator Assemblages in the Florida Everglades.Appl Environ Microbiol. 2019 Jun 17;85(13):e00156-19. doi: 10.1128/AEM.00156-19. Print 2019 Jul 1. Appl Environ Microbiol. 2019. PMID: 31028023 Free PMC article.
-
Determining the Reliability of Measuring Mercury Cycling Gene Abundance with Correlations with Mercury and Methylmercury Concentrations.Environ Sci Technol. 2019 Aug 6;53(15):8649-8663. doi: 10.1021/acs.est.8b06389. Epub 2019 Jul 12. Environ Sci Technol. 2019. PMID: 31260289
-
Microbial Mercury Methylation in Aquatic Environments: A Critical Review of Published Field and Laboratory Studies.Environ Sci Technol. 2019 Jan 2;53(1):4-19. doi: 10.1021/acs.est.8b02709. Epub 2018 Dec 21. Environ Sci Technol. 2019. PMID: 30525497 Review.
-
Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: a critical review.Environ Sci Technol. 2013 Mar 19;47(6):2441-56. doi: 10.1021/es304370g. Epub 2013 Feb 27. Environ Sci Technol. 2013. PMID: 23384298 Review.
Cited by
-
Robust Mercury Methylation across Diverse Methanogenic Archaea.mBio. 2018 Apr 10;9(2):e02403-17. doi: 10.1128/mBio.02403-17. mBio. 2018. PMID: 29636434 Free PMC article.
-
Overview of Methylation and Demethylation Mechanisms and Influencing Factors of Mercury in Water.Toxics. 2024 Sep 30;12(10):715. doi: 10.3390/toxics12100715. Toxics. 2024. PMID: 39453135 Free PMC article. Review.
-
Mercury-methylating bacteria are associated with copepods: A proof-of-principle survey in the Baltic Sea.PLoS One. 2020 Mar 16;15(3):e0230310. doi: 10.1371/journal.pone.0230310. eCollection 2020. PLoS One. 2020. PMID: 32176728 Free PMC article.
-
Effects of Organic Phosphorus on Methylotrophic Methanogenesis in Coastal Lagoon Sediments With Seagrass (Zostera marina) Colonization.Front Microbiol. 2020 Jul 31;11:1770. doi: 10.3389/fmicb.2020.01770. eCollection 2020. Front Microbiol. 2020. PMID: 32849394 Free PMC article.
-
Nutrient Exposure Alters Microbial Composition, Structure, and Mercury Methylating Activity in Periphyton in a Contaminated Watershed.Front Microbiol. 2021 Mar 19;12:647861. doi: 10.3389/fmicb.2021.647861. eCollection 2021. Front Microbiol. 2021. PMID: 33815336 Free PMC article.
References
-
- Marvin-DiPasquale M, Windham-Myers L, Agee JL, Kakouros E, Kieu LH, Fleck JA, Alpers CN, Stricker CA. 2014. Methylmercury production in sediment from agricultural and non-agricultural wetlands in the Yolo Bypass, California, USA. Sci Tot Environ 484:288–299. doi:10.1016/j.scitotenv.2013.09.098. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

