A Genetic System for the Thermophilic Acetogenic Bacterium Thermoanaerobacter kivui
- PMID: 29150512
- PMCID: PMC5772241
- DOI: 10.1128/AEM.02210-17
A Genetic System for the Thermophilic Acetogenic Bacterium Thermoanaerobacter kivui
Abstract
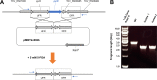
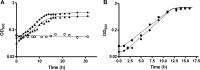
Thermoanaerobacter kivui is one of the very few thermophilic acetogenic microorganisms. It grows optimally at 66°C on sugars but also lithotrophically with H2 + CO2 or with CO, producing acetate as the major product. While a genome-derived model of acetogenesis has been developed, only a few physiological or biochemical experiments regarding the function of important enzymes in carbon and energy metabolism have been carried out. To address this issue, we developed a method for targeted markerless gene deletions and for integration of genes into the genome of T. kivui The strain naturally took up plasmid DNA in the exponential growth phase, with a transformation frequency of up to 3.9 × 10-6 A nonreplicating plasmid and selection with 5-fluoroorotate was used to delete the gene encoding the orotate phosphoribosyltransferase (pyrE), resulting in a ΔpyrE uracil-auxotrophic strain, TKV002. Reintroduction of pyrE on a plasmid or insertion of pyrE into different loci within the genome restored growth without uracil. We subsequently studied fructose metabolism in T. kivui The gene fruK (TKV_c23150) encoding 1-phosphofructosekinase (1-PFK) was deleted, using pyrE as a selective marker via two single homologous recombination events. The resulting ΔfruK strain, TKV003, did not grow on fructose; however, growth on glucose (or on mannose) was unaffected. The combination of pyrE as a selective marker and the natural competence of the strain for DNA uptake will be the basis for future studies on CO2 reduction and energy conservation and their regulation in this thermophilic acetogenic bacterium.IMPORTANCE Acetogenic bacteria are currently the focus of research toward biotechnological applications due to their potential for de novo synthesis of carbon compounds such as acetate, butyrate, or ethanol from H2 + CO2 or from synthesis gas. Based on available genome sequences and on biochemical experiments, acetogens differ in their energy metabolism. Thus, there is an urgent need to understand the carbon and electron flows through the Wood-Ljungdahl pathway and their links to energy conservation, which requires genetic manipulations such as deletion or overexpression of genes encoding putative key enzymes. Unfortunately, genetic systems have been reported for only a few acetogenic bacteria. Here, we demonstrate proof of concept for the genetic modification of the thermophilic acetogenic species Thermoanaerobacter kivui The genetic system will be used to study genes involved in biosynthesis and energy metabolism, and may further be applied to metabolically engineer T. kivui to produce fuels and chemicals.
Keywords: DNA uptake; Thermoanaerobacter kivui; acetogenesis; fructose metabolism; genetic system.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
The energy-converting hydrogenase Ech2 is important for the growth of the thermophilic acetogen Thermoanaerobacter kivui on ferredoxin-dependent substrates.Microbiol Spectr. 2024 Apr 2;12(4):e0338023. doi: 10.1128/spectrum.03380-23. Epub 2024 Feb 22. Microbiol Spectr. 2024. PMID: 38385688 Free PMC article.
-
A genome-guided analysis of energy conservation in the thermophilic, cytochrome-free acetogenic bacterium Thermoanaerobacter kivui.BMC Genomics. 2014 Dec 18;15(1):1139. doi: 10.1186/1471-2164-15-1139. BMC Genomics. 2014. PMID: 25523312 Free PMC article.
-
A thermostable mannitol-1-phosphate dehydrogenase is required in mannitol metabolism of the thermophilic acetogenic bacterium Thermoanaerobacter kivui.Environ Microbiol. 2019 Oct;21(10):3728-3736. doi: 10.1111/1462-2920.14720. Epub 2019 Jul 10. Environ Microbiol. 2019. PMID: 31219674
-
"Hot" acetogenesis.Extremophiles. 2017 Jan;21(1):15-26. doi: 10.1007/s00792-016-0873-3. Epub 2016 Sep 13. Extremophiles. 2017. PMID: 27623994 Review.
-
New Horizons in Acetogenic Conversion of One-Carbon Substrates and Biological Hydrogen Storage.Trends Biotechnol. 2019 Dec;37(12):1344-1354. doi: 10.1016/j.tibtech.2019.05.008. Epub 2019 Jun 27. Trends Biotechnol. 2019. PMID: 31257058 Review.
Cited by
-
The monofunctional CO dehydrogenase CooS is essential for growth of Thermoanaerobacter kivui on carbon monoxide.Extremophiles. 2021 Dec 17;26(1):4. doi: 10.1007/s00792-021-01251-y. Extremophiles. 2021. PMID: 34919167 Free PMC article.
-
Synthetic Biology on Acetogenic Bacteria for Highly Efficient Conversion of C1 Gases to Biochemicals.Int J Mol Sci. 2020 Oct 15;21(20):7639. doi: 10.3390/ijms21207639. Int J Mol Sci. 2020. PMID: 33076477 Free PMC article. Review.
-
The Rnf Complex Is an Energy-Coupled Transhydrogenase Essential To Reversibly Link Cellular NADH and Ferredoxin Pools in the Acetogen Acetobacterium woodii.J Bacteriol. 2018 Oct 10;200(21):e00357-18. doi: 10.1128/JB.00357-18. Print 2018 Nov 1. J Bacteriol. 2018. PMID: 30126940 Free PMC article.
-
The pyruvate:ferredoxin oxidoreductase of the thermophilic acetogen, Thermoanaerobacter kivui.FEBS Open Bio. 2021 May;11(5):1332-1342. doi: 10.1002/2211-5463.13136. Epub 2021 Apr 4. FEBS Open Bio. 2021. PMID: 33660937 Free PMC article.
-
The energy-converting hydrogenase Ech2 is important for the growth of the thermophilic acetogen Thermoanaerobacter kivui on ferredoxin-dependent substrates.Microbiol Spectr. 2024 Apr 2;12(4):e0338023. doi: 10.1128/spectrum.03380-23. Epub 2024 Feb 22. Microbiol Spectr. 2024. PMID: 38385688 Free PMC article.
References
-
- Schink B, Stams A. 2006. Syntrophism among prokaryotes, p 309–336. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed, vol 2 Springer Science+Business Media, Berlin, Germany.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

