ROP GTPases Structure-Function and Signaling Pathways
- PMID: 29150557
- PMCID: PMC5761820
- DOI: 10.1104/pp.17.01415
ROP GTPases Structure-Function and Signaling Pathways
Abstract
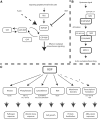
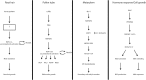
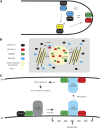
Interactions between receptor like kinases and guanyl nucleotide exchange factors together with identification of effector proteins reveal putative ROP GTPases signaling cascades.
Figures






Similar articles
-
The cellular function of ROP GTPase prenylation is important for multicellularity in the moss Physcomitrium patens.Development. 2022 Jun 15;149(12):dev200279. doi: 10.1242/dev.200279. Epub 2022 Jun 23. Development. 2022. PMID: 35660859
-
Small GTPases in vesicle trafficking.Curr Opin Plant Biol. 2004 Dec;7(6):694-700. doi: 10.1016/j.pbi.2004.09.014. Curr Opin Plant Biol. 2004. PMID: 15491918 Review.
-
ROP/RAC GTPase: an old new master regulator for plant signaling.Curr Opin Plant Biol. 2004 Oct;7(5):527-36. doi: 10.1016/j.pbi.2004.07.006. Curr Opin Plant Biol. 2004. PMID: 15337095 Review.
-
GTPases: a family of molecular switches and clocks.Philos Trans R Soc Lond B Biol Sci. 1995 Sep 29;349(1329):283-9. doi: 10.1098/rstb.1995.0114. Philos Trans R Soc Lond B Biol Sci. 1995. PMID: 8577839 Review.
-
ROP/RAC GTPase signaling.Curr Opin Plant Biol. 2007 Oct;10(5):490-4. doi: 10.1016/j.pbi.2007.07.005. Epub 2007 Aug 20. Curr Opin Plant Biol. 2007. PMID: 17709276 Free PMC article. Review.
Cited by
-
Microtubule-associated phase separation of MIDD1 tunes cell wall spacing in xylem vessels in Arabidopsis thaliana.Nat Plants. 2024 Jan;10(1):100-117. doi: 10.1038/s41477-023-01593-9. Epub 2024 Jan 3. Nat Plants. 2024. PMID: 38172572
-
Capsicum Leaves under Stress: Using Multi-Omics Analysis to Detect Abiotic Stress Network of Secondary Metabolism in Two Species.Antioxidants (Basel). 2022 Mar 30;11(4):671. doi: 10.3390/antiox11040671. Antioxidants (Basel). 2022. PMID: 35453356 Free PMC article.
-
ZmRop1 participates in maize defense response to the damage of Spodoptera frugiperda larvae through mediating ROS and soluble phenol production.Plant Direct. 2022 Dec 15;6(12):e468. doi: 10.1002/pld3.468. eCollection 2022 Dec. Plant Direct. 2022. PMID: 36540415 Free PMC article.
-
Abscisic acid switches cell division modes of asymmetric cell division and symmetric cell division in stem cells of protonemal filaments in the moss Physcomitrium patens.Plant Biotechnol (Tokyo). 2022 Mar 25;39(1):13-17. doi: 10.5511/plantbiotechnology.22.0107a. Plant Biotechnol (Tokyo). 2022. PMID: 35800966 Free PMC article.
-
The Arabidopsis Rho of Plants GTPase ROP1 Is a Potential Calcium-Dependent Protein Kinase (CDPK) Substrate.Plants (Basel). 2021 Sep 29;10(10):2053. doi: 10.3390/plants10102053. Plants (Basel). 2021. PMID: 34685862 Free PMC article.
References
-
- Abo A, Pick E, Hall A, Totty N, Teahan CG, Segal AW (1991) Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature 353: 668–670 - PubMed
-
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, et al. (2013) An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476 - PubMed
-
- Barbosa IC, Zourelidou M, Willige BC, Weller B, Schwechheimer C (2014) D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev Cell 29: 674–685 - PubMed
-
- Basu D, El-Assal Sel-D, Le J, Mallery EL, Szymanski DB (2004) Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131: 4345–4355 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

