Mapping tenascin-C interaction with toll-like receptor 4 reveals a new subset of endogenous inflammatory triggers
- PMID: 29150600
- PMCID: PMC5693923
- DOI: 10.1038/s41467-017-01718-7
Mapping tenascin-C interaction with toll-like receptor 4 reveals a new subset of endogenous inflammatory triggers
Abstract
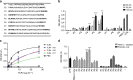
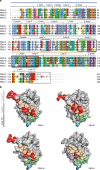
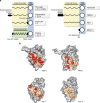
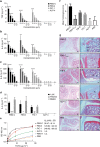
Pattern recognition underpins innate immunity; the accurate identification of danger, including infection, injury, or tumor, is key to an appropriately targeted immune response. Pathogen detection is increasingly well defined mechanistically, but the discrimination of endogenous inflammatory triggers remains unclear. Tenascin-C, a matrix protein induced upon tissue damage and expressed by tumors, activates toll-like receptor 4 (TLR4)-mediated sterile inflammation. Here we map three sites within tenascin-C that directly and cooperatively interact with TLR4. We also identify a conserved inflammatory epitope in related proteins from diverse families, and demonstrate that its presence targets molecules for TLR detection, while its absence enables escape of innate immune surveillance. These data reveal a unique molecular code that defines endogenous proteins as inflammatory stimuli by marking them for recognition by TLRs.
Conflict of interest statement
K.S.M. is the founder of, and consultant to, Nascient Ltd. The remaining authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

