Genome-wide survey indicates diverse physiological roles of the turnip (Brassica rapa var. rapa) calcium-dependent protein kinase genes
- PMID: 29150669
- PMCID: PMC5693941
- DOI: 10.1038/s41598-017-16102-0
Genome-wide survey indicates diverse physiological roles of the turnip (Brassica rapa var. rapa) calcium-dependent protein kinase genes
Abstract
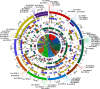
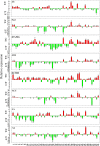
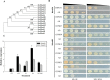
Calcium-dependent protein kinases (CDPKs) as crucial sensors of calcium concentration changes play important roles in responding to abiotic and biotic stresses. In this study, 55 BrrCDPK genes, which were phylogenetically clustered into four subfamilies, were identified. Chromosome locations indicated that the CDPK family in turnip expanded by segmental duplication and genome rearrangement. Moreover, gene expression profiles showed that different BrrCDPKs were expressed in specific tissues or stages. Transcript levels of BrrCDPKs indicated that they were involved in abiotic and biotic stresses and that paralogs exhibited functional divergence. Additionally, we identified 15 Rboh genes in turnip; the results of yeast two-hybrid analysis suggested that BrrRbohD1 interacted only with BrrCDPK10 and that BrrRbohD2 interacted with BrrCDPK4/7/9/10/17/22/23. Most of the genes play an important role in pst DC3000 defense by regulating the accumulation of H2O2 and stomatal closure. Our study may provide an important foundation for future functional analysis of BrrCDPKs and reveal further biological roles.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Genome wide survey, evolution and expression analysis of PHD finger genes reveal their diverse roles during the development and abiotic stress responses in Brassica rapa L.BMC Genomics. 2019 Oct 24;20(1):773. doi: 10.1186/s12864-019-6080-8. BMC Genomics. 2019. PMID: 31651238 Free PMC article.
-
Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapa ssp. pekinensis).BMC Genomics. 2017 Nov 16;18(1):885. doi: 10.1186/s12864-017-4277-2. BMC Genomics. 2017. PMID: 29145809 Free PMC article.
-
Functional Differentiation of the Duplicated Gene BrrCIPK9 in Turnip (Brassica rapa var. rapa).Genes (Basel). 2024 Mar 26;15(4):405. doi: 10.3390/genes15040405. Genes (Basel). 2024. PMID: 38674340 Free PMC article.
-
Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa.Mol Genet Genomics. 2015 Feb;290(1):79-95. doi: 10.1007/s00438-014-0898-1. Epub 2014 Aug 23. Mol Genet Genomics. 2015. PMID: 25149146
-
Advances in Protein Kinase Regulation of Stress Responses in Fruits and Vegetables.Int J Mol Sci. 2025 Jan 17;26(2):768. doi: 10.3390/ijms26020768. Int J Mol Sci. 2025. PMID: 39859482 Free PMC article. Review.
Cited by
-
Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in grass Brachypodium distachyon.BMC Genomics. 2020 Jan 16;21(1):53. doi: 10.1186/s12864-020-6475-6. BMC Genomics. 2020. PMID: 31948407 Free PMC article.
-
The Interaction Between StCDPK14 and StRbohB Contributes to Benzo-(1, 2, 3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester-Induced Wound Healing of Potato Tubers by Regulating Reactive Oxygen Species Generation.Front Plant Sci. 2021 Nov 15;12:737524. doi: 10.3389/fpls.2021.737524. eCollection 2021. Front Plant Sci. 2021. PMID: 34868121 Free PMC article.
-
Genome-Wide Identification and Expression Profile Analysis of the NADPH Oxidase Gene Family in Avena sativa L.Int J Mol Sci. 2025 Mar 13;26(6):2576. doi: 10.3390/ijms26062576. Int J Mol Sci. 2025. PMID: 40141217 Free PMC article.
-
Genome-wide identification of CDPKs in jujube and expression profile analysis in response to multiple biological processes.Front Plant Sci. 2025 May 27;16:1593446. doi: 10.3389/fpls.2025.1593446. eCollection 2025. Front Plant Sci. 2025. PMID: 40496467 Free PMC article.
-
Genome-wide survey of Calcium-Dependent Protein Kinases (CPKs) in five Brassica species and identification of CPKs induced by Plasmodiophora brassicae in B. rapa, B. oleracea, and B. napus.Front Plant Sci. 2022 Nov 21;13:1067723. doi: 10.3389/fpls.2022.1067723. eCollection 2022. Front Plant Sci. 2022. PMID: 36479517 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

