cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate
- PMID: 29150937
- PMCID: PMC5852640
- DOI: 10.1002/bit.26498
cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate
Abstract
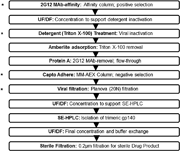
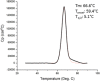
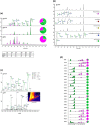
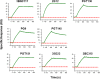
We describe the properties of BG505 SOSIP.664 HIV-1 envelope glycoprotein trimers produced under current Good Manufacturing Practice (cGMP) conditions. These proteins are the first of a new generation of native-like trimers that are the basis for many structure-guided immunogen development programs aimed at devising how to induce broadly neutralizing antibodies (bNAbs) to HIV-1 by vaccination. The successful translation of this prototype demonstrates the feasibility of producing similar immunogens on an appropriate scale and of an acceptable quality for Phase I experimental medicine clinical trials. BG505 SOSIP.664 trimers are extensively glycosylated, contain numerous disulfide bonds and require proteolytic cleavage, all properties that pose a substantial challenge to cGMP production. Our strategy involved creating a stable CHO cell line that was adapted to serum-free culture conditions to produce envelope glycoproteins. The trimers were then purified by chromatographic methods using a 2G12 bNAb affinity column and size-exclusion chromatography. The chosen procedures allowed any adventitious viruses to be cleared from the final product to the required extent of >12 log10 . The final cGMP production run yielded 3.52 g (peptidic mass) of fully purified trimers (Drug Substance) from a 200 L bioreactor, a notable yield for such a complex glycoprotein. The purified trimers were fully native-like as judged by negative-stain electron microscopy, and were stable over a multi-month period at room temperature or below and for at least 1 week at 50°C. Their antigenicity, disulfide bond patterns, and glycan composition were consistent with trimers produced on a research laboratory scale. The methods reported here should pave the way for the cGMP production of other native-like Env glycoprotein trimers of various designs and genotypes.
Keywords: HIV-1 vaccine; SOSIP; affinity purification; cGMP; native-like Env trimers; recombinant vaccine development.
© 2017 The Authors. Biotechnology and Bioengineering Published by Wiley Periodicals, Inc.
Figures

References
-
- Bleck, G. T. 2010. GPEx® A flexible method for the rapid generation of stable, high expressing, antibody producing mammalian cell lines. American Association of Pharmaceutical Scientists, 12.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

