Mammalian mitophagy - from in vitro molecules to in vivo models
- PMID: 29151277
- PMCID: PMC5947125
- DOI: 10.1111/febs.14336
Mammalian mitophagy - from in vitro molecules to in vivo models
Abstract
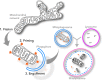
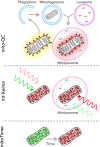
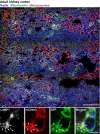
The autophagic turnover of mitochondria, termed mitophagy, is thought to play an essential role in not only maintaining the health of the mitochondrial network but also that of the cell and organism as a whole. We have come a long way in identifying the molecular components required for mitophagy through extensive in vitro work and cell line characterisation, yet the physiological significance and context of these pathways remain largely unexplored. This is highlighted by the recent development of new mouse models that have revealed a striking level of variation in mitophagy, even under normal conditions. Here, we focus on programmed mitophagy and summarise our current understanding of why, how and where this takes place in mammals.
Keywords: NIX; Parkin; autophagy; development; disease; metabolism; mito-QC; mitochondria; mitophagy; mouse models.
© 2017 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures
References
-
- Ney PA (2015) Mitochondrial autophagy: origins, significance, and role of BNIP3 and NIX. Biochim Biophys Acta 1853, 2775–2783. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

