Identifying and analyzing different cancer subtypes using RNA-seq data of blood platelets
- PMID: 29152097
- PMCID: PMC5675649
- DOI: 10.18632/oncotarget.20903
Identifying and analyzing different cancer subtypes using RNA-seq data of blood platelets
Abstract
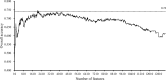
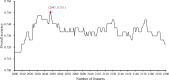
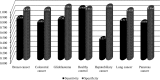
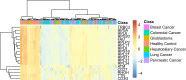
Detection and diagnosis of cancer are especially important for early prevention and effective treatments. Traditional methods of cancer detection are usually time-consuming and expensive. Liquid biopsy, a newly proposed noninvasive detection approach, can promote the accuracy and decrease the cost of detection according to a personalized expression profile. However, few studies have been performed to analyze this type of data, which can promote more effective methods for detection of different cancer subtypes. In this study, we applied some reliable machine learning algorithms to analyze data retrieved from patients who had one of six cancer subtypes (breast cancer, colorectal cancer, glioblastoma, hepatobiliary cancer, lung cancer and pancreatic cancer) as well as healthy persons. Quantitative gene expression profiles were used to encode each sample. Then, they were analyzed by the maximum relevance minimum redundancy method. Two feature lists were obtained in which genes were ranked rigorously. The incremental feature selection method was applied to the mRMR feature list to extract the optimal feature subset, which can be used in the support vector machine algorithm to determine the best performance for the detection of cancer subtypes and healthy controls. The ten-fold cross-validation for the constructed optimal classification model yielded an overall accuracy of 0.751. On the other hand, we extracted the top eighteen features (genes), including TTN, RHOH, RPS20, TRBC2, in another feature list, the MaxRel feature list, and performed a detailed analysis of them. The results indicated that these genes could be important biomarkers for discriminating different cancer subtypes and healthy controls.
Keywords: RNA-seq data; cancer detection; liquid biopsy; maximum relevance minimum redundancy; support vector machine.
Conflict of interest statement
CONFLICTS OF INTEREST No potential conflicts of interest were disclosed.
Figures

Similar articles
-
Identification and Analysis of Glioblastoma Biomarkers Based on Single Cell Sequencing.Front Bioeng Biotechnol. 2020 Mar 5;8:167. doi: 10.3389/fbioe.2020.00167. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32195242 Free PMC article.
-
mRMR-ABC: A Hybrid Gene Selection Algorithm for Cancer Classification Using Microarray Gene Expression Profiling.Biomed Res Int. 2015;2015:604910. doi: 10.1155/2015/604910. Epub 2015 Apr 15. Biomed Res Int. 2015. PMID: 25961028 Free PMC article.
-
Computational method for distinguishing lysine acetylation, sumoylation, and ubiquitination using the random forest algorithm with a feature selection procedure.Comb Chem High Throughput Screen. 2017 Dec 17. doi: 10.2174/1386207321666171218114056. Online ahead of print. Comb Chem High Throughput Screen. 2017. PMID: 29256343
-
The feature selection bias problem in relation to high-dimensional gene data.Artif Intell Med. 2016 Jan;66:63-71. doi: 10.1016/j.artmed.2015.11.001. Epub 2015 Nov 14. Artif Intell Med. 2016. PMID: 26674595
-
Identification of biomarkers for hepatocellular carcinoma based on single cell sequencing and machine learning algorithms.Front Genet. 2022 Oct 24;13:873218. doi: 10.3389/fgene.2022.873218. eCollection 2022. Front Genet. 2022. PMID: 36353113 Free PMC article.
Cited by
-
Is There a Role for Machine Learning in Liquid Biopsy for Brain Tumors? A Systematic Review.Int J Mol Sci. 2023 Jun 3;24(11):9723. doi: 10.3390/ijms24119723. Int J Mol Sci. 2023. PMID: 37298673 Free PMC article.
-
TriCLFF: a multi-modal feature fusion framework using contrastive learning for spatial domain identification.Brief Bioinform. 2025 Jul 2;26(4):bbaf316. doi: 10.1093/bib/bbaf316. Brief Bioinform. 2025. PMID: 40639417 Free PMC article.
-
Bioinformatic profiling identifies prognosis-related genes in the immune microenvironment of endometrial carcinoma.Sci Rep. 2021 Jun 15;11(1):12608. doi: 10.1038/s41598-021-92091-5. Sci Rep. 2021. PMID: 34131259 Free PMC article.
-
Identification and Analysis of Dysfunctional Genes and Pathways in CD8+ T Cells of Non-Small Cell Lung Cancer Based on RNA Sequencing.Front Genet. 2020 May 8;11:352. doi: 10.3389/fgene.2020.00352. eCollection 2020. Front Genet. 2020. PMID: 32457792 Free PMC article.
-
Assessing the Influence of Climate Change and Environmental Factors on the Top Tick-Borne Diseases in the United States: A Systematic Review.Microorganisms. 2023 Dec 27;12(1):50. doi: 10.3390/microorganisms12010050. Microorganisms. 2023. PMID: 38257877 Free PMC article. Review.
References
-
- Krishnan A, Nair SA, Pillai MR. Biology of PPAR gamma in cancer: A critical review on existing lacunae. Current molecular medicine. 2007;7:532–540. - PubMed
-
- Carney DN. The Biology Of Lung-Cancer - a Review. Acta Oncol. 1989;28:1–5. - PubMed
-
- Shaw P, Costa J. Molecular-Biology Of Colon Cancer - (Review) Anticancer Research. 1989;9:21–27. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

