Breast tissue, oral and urinary microbiomes in breast cancer
- PMID: 29152146
- PMCID: PMC5675698
- DOI: 10.18632/oncotarget.21490
Breast tissue, oral and urinary microbiomes in breast cancer
Abstract
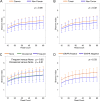
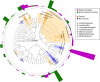
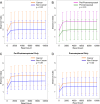
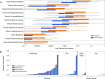
It has long been proposed that the gut microbiome contributes to breast carcinogenesis by modifying systemic estrogen levels. This is often cited as a possible mechanism linking breast cancer and high-fat, low-fiber diets as well as antibiotic exposure, associations previously identified in population-based studies. More recently, a distinct microbiome has been identified within breast milk and tissue, but few studies have characterized differences in the breast tissue microbiota of patients with and without cancer, and none have investigated distant body-site microbiomes outside of the gut. We hypothesize that cancerous breast tissue is associated with a microbiomic profile distinct from that of benign breast tissue, and that microbiomes of more distant sites, the oral cavity and urinary tract, will reflect dysbiosis as well. Fifty-seven women with invasive breast cancer undergoing mastectomy and 21 healthy women undergoing cosmetic breast surgery were enrolled. The bacterial 16S rRNA gene was amplified from urine, oral rinse and surgically collected breast tissue, sequenced, and processed through a QIIME-based bioinformatics pipeline. Cancer patient breast tissue microbiomes clustered significantly differently from non-cancer patients (p=0.03), largely driven by decreased relative abundance of Methylobacterium in cancer patients (median 0.10 vs. 0.24, p=0.03). There were no significant differences in oral rinse samples. Differences in urinary microbiomes were largely explained by menopausal status, with peri/postmenopausal women showing decreased levels of Lactobacillus. Independent of menopausal status, however, cancer patients had increased levels of gram-positive organisms including Corynebacterium (p<0.01), Staphylococcus (p=0.02), Actinomyces (p<0.01), and Propionibacteriaceae (p<0.01). Our observations suggest that the local breast microbiota differ in patients with and without breast cancer. Cancer patient urinary microbiomes were characterized by increased levels of gram-positive organisms in this study, but need to be further studied in larger cohorts.
Keywords: breast cancer; metagenomics; microbiome; oral; urine.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that they have no relevant conflicts of interest.
Figures




References
-
- World Health Organization World Health Statistics 2012 WHO. 2012. p. 171.
-
- Lacey JV, Kreimer AR, Buys SS, Marcus PM, Chang SC, Leitzmann MF, Hoover RN, Prorok PC, Berg CD, Hartge P. Breast cancer epidemiology according to recognized breast cancer risk factors in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial Cohort. BMC Cancer. 2009;9:84. https://doi.org/10.1186/1471-2407-9-84 - DOI - PMC - PubMed
-
- Madigan MP, Ziegler RG, Benichou J, Byrne C, Hoover RN. Proportion of breast cancer cases in the United States explained by well-established risk factors. J Natl Cancer Inst. 1995;87:1681–5. https://doi.org/10.1093/jnci/87.22.1681 - DOI - PubMed
-
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. https://doi.org/10.1101/gr.096651.109 - DOI - PMC - PubMed
-
- Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35:249–55. https://doi.org/10.1093/carcin/bgt392 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

