Evaluation and optimization of microbial DNA extraction from fecal samples of wild Antarctic bird species
- PMID: 29152162
- PMCID: PMC5678435
- DOI: 10.1080/20008686.2017.1386536
Evaluation and optimization of microbial DNA extraction from fecal samples of wild Antarctic bird species
Abstract
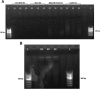
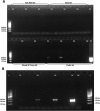
Introduction: Advances in the development of nucleic acid-based methods have dramatically facilitated studies of host-microbial interactions. Fecal DNA analysis can provide information about the host's microbiota and gastrointestinal pathogen burden. Numerous studies have been conducted in mammals, yet birds are less well studied. Avian fecal DNA extraction has proved challenging, partly due to the mixture of fecal and urinary excretions and the deficiency of optimized protocols. This study presents an evaluation of the performance in avian fecal DNA extraction of six commercial kits from different bird species, focusing on penguins. Material and methods: Six DNA extraction kits were first tested according to the manufacturers' instructions using mallard feces. The kit giving the highest DNA yield was selected for further optimization and evaluation using Antarctic bird feces. Results: Penguin feces constitute a challenging sample type: most of the DNA extraction kits failed to yield acceptable amounts of DNA. The QIAamp cador Pathogen kit (Qiagen) performed the best in the initial investigation. Further optimization of the protocol resulted in good yields of high-quality DNA from seven bird species of different avian orders. Conclusion: This study presents an optimized approach to DNA extraction from challenging avian fecal samples.
Keywords: Antarctica; Aves; DNA extraction; feces; method evaluation and scatology.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
The choice of DNA extraction protocol affects the quantification of gut microbiomes in two passerines.J Microbiol Methods. 2025 Aug;235:107144. doi: 10.1016/j.mimet.2025.107144. Epub 2025 May 8. J Microbiol Methods. 2025. PMID: 40345500
-
Microbial diversity in fecal samples depends on DNA extraction method: easyMag DNA extraction compared to QIAamp DNA stool mini kit extraction.BMC Res Notes. 2014 Jan 21;7:50. doi: 10.1186/1756-0500-7-50. BMC Res Notes. 2014. PMID: 24447346 Free PMC article.
-
Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens.Nutr J. 2010 May 22;9:23. doi: 10.1186/1475-2891-9-23. Nutr J. 2010. PMID: 20492702 Free PMC article.
-
Evaluation of fecal DNA extraction protocols for human gut microbiome studies.BMC Microbiol. 2020 Jul 17;20(1):212. doi: 10.1186/s12866-020-01894-5. BMC Microbiol. 2020. PMID: 32680572 Free PMC article.
-
Comparison of the techniques for isolating immunoassay-suitable proteins from heterogeneous fecal samples.Anal Biochem. 2025 Mar;698:115748. doi: 10.1016/j.ab.2024.115748. Epub 2024 Dec 10. Anal Biochem. 2025. PMID: 39667549 Review.
Cited by
-
The impact of management on the fecal microbiome of endangered greater sage-grouse (Centrocercus urophasianus) in a zoo-based conservation program.Conserv Physiol. 2024 Aug 7;12(1):coae052. doi: 10.1093/conphys/coae052. eCollection 2024. Conserv Physiol. 2024. PMID: 39113731 Free PMC article.
-
The promise and challenge of cancer microbiome research.Genome Biol. 2020 Jun 2;21(1):131. doi: 10.1186/s13059-020-02037-9. Genome Biol. 2020. PMID: 32487228 Free PMC article. Review.
-
Alteration of the Gut Microbiome in Normal and Overweight School Children from Selangor with Lactobacillus Fermented Milk Administration.Evol Bioinform Online. 2020 Nov 23;16:1176934320965943. doi: 10.1177/1176934320965943. eCollection 2020. Evol Bioinform Online. 2020. PMID: 33281440 Free PMC article.
-
Curing piglets from diarrhea and preparation of a healthy microbiome with Bacillus treatment for industrial animal breeding.Sci Rep. 2020 Nov 10;10(1):19476. doi: 10.1038/s41598-020-75207-1. Sci Rep. 2020. PMID: 33173074 Free PMC article.
-
Gut microbiome is affected by inter-sexual and inter-seasonal variation in diet for thick-billed murres (Uria lomvia).Sci Rep. 2021 Jan 13;11(1):1200. doi: 10.1038/s41598-020-80557-x. Sci Rep. 2021. PMID: 33441848 Free PMC article.
References
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–9. - PubMed
-
- Fraher MH, O’Toole PW, Quigley EMM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9:312–322. - PubMed
-
- Kelly CD. IV. Bacteria. Bot Rev. 1954;20:417–424.
-
- Levin M. Les microbes dans les regions arctiques. Ann Inst Pasteur. 1899;13:558–567.
LinkOut - more resources
Full Text Sources
Other Literature Sources
