Identification of Equid herpesvirus 2 in tissue-engineered equine tendon
- PMID: 29152595
- PMCID: PMC5664983
- DOI: 10.12688/wellcomeopenres.12176.2
Identification of Equid herpesvirus 2 in tissue-engineered equine tendon
Abstract
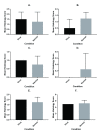
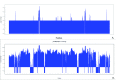
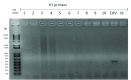
Background: Incidental findings of virus-like particles were identified following electron microscopy of tissue-engineered tendon constructs (TETC) derived from equine tenocytes. We set out to determine the nature of these particles, as there are few studies which identify virus in tendons per se, and their presence could have implications for tissue-engineering using allogenic grafts. Methods: Virus particles were identified in electron microscopy of TETCs. Virion morphology was used to initially hypothesise the virus identity. Next generation sequencing was implemented to identify the virus. A pan herpesvirus PCR was used to validate the RNASeq findings using an independent platform. Histological analysis and biochemical analysis was undertaken on the TETCs. Results: Morphological features suggested the virus to be either a retrovirus or herpesvirus. Subsequent next generation sequencing mapped reads to Equid herpesvirus 2 (EHV2). Histological examination and biochemical testing for collagen content revealed no significant differences between virally affected TETCs and non-affected TETCs. An independent set of equine superficial digital flexor tendon tissue (n=10) examined using designed primers for specific EHV2 contigs identified at sequencing were negative. These data suggest that EHV is resident in some equine tendon. Conclusions: EHV2 was demonstrated in equine tenocytes for the first time; likely from in vivo infection. The presence of EHV2 could have implications to both tissue-engineering and tendinopathy.
Keywords: Equine herpesvirus 2; equine; next-generation sequencing; superficial digital flexor tendon; tissue-engineered tendon.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

