APOBEC Enzymes as Targets for Virus and Cancer Therapy
- PMID: 29153851
- PMCID: PMC5775913
- DOI: 10.1016/j.chembiol.2017.10.007
APOBEC Enzymes as Targets for Virus and Cancer Therapy
Abstract
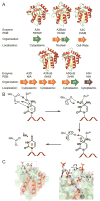
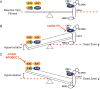
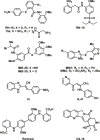
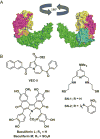
Human DNA cytosine-to-uracil deaminases catalyze mutations in both pathogen and cellular genomes. APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H restrict human immunodeficiency virus 1 (HIV-1) infection in cells deficient in the viral infectivity factor (Vif), and have the potential to catalyze sublethal levels of mutation in viral genomes in Vif-proficient cells. At least two APOBEC3 enzymes, and in particular APOBEC3B, are sources of somatic mutagenesis in cancer cells that drive tumor evolution and may manifest clinically as recurrence, metastasis, and/or therapy resistance. Consequently, APOBEC3 enzymes are tantalizing targets for developing chemical probes and therapeutic molecules to harness mutational processes in human disease. This review highlights recent efforts to chemically manipulate APOBEC3 activities.
Keywords: APOBEC; cancer; chemical probes; cytosine-to-uracil; mutation; virus.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

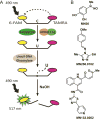
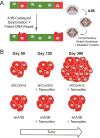
References
-
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. - PubMed
-
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

